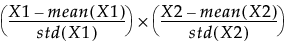|
•
|
lignin sulfonate (ls), which is pulp industry pollution
|
|
•
|
humic acid (ha), which is a natural forest product
|
|
•
|
|
1.
|
Note: The data in the Baltic.jmp data table are reported in Umetrics (1995). The original source is Lindberg, Persson, and Wold (1983).
|
2.
|
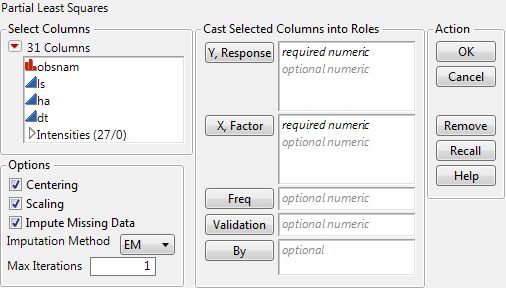
Select Analyze > Multivariate Methods > Partial Least Squares.
|
|
3.
|
|
4.
|
Assign Intensities, which contains the 27 intensity variables v1 through v27, to the X, Factor role.
|
|
5.
|
Click OK.
|
|
6.
|
Select Leave-One-Out as the Validation Method.
|
|
7.
|
Click Go.
|
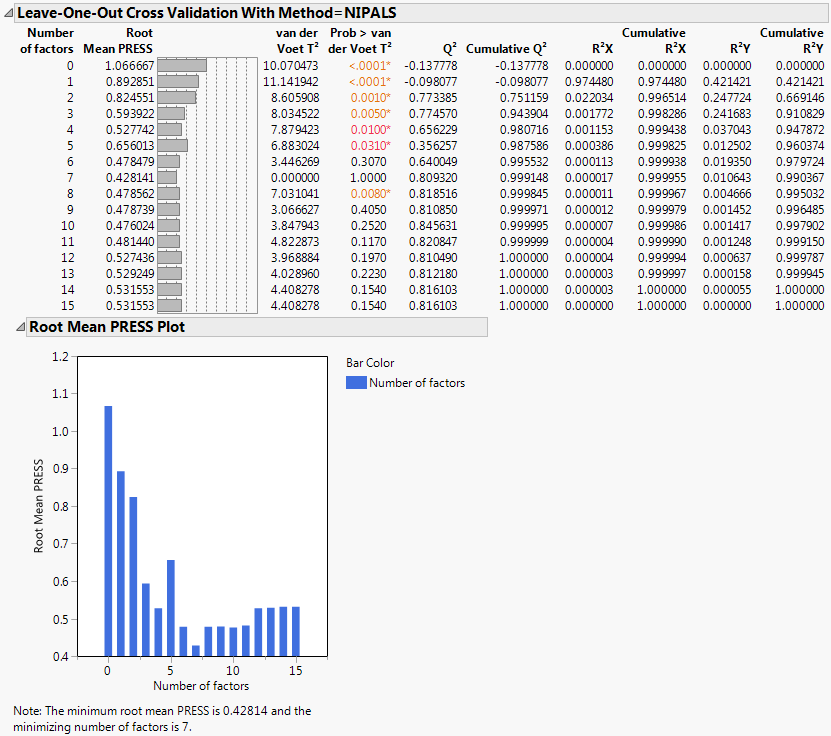
A portion of the report appears in Partial Least Squares Report. Since the van der Voet test is a randomization test, your Prob > van der Voet T2 values can differ slightly from those in Partial Least Squares Report.
The Root Mean PRESS (predicted residual sum of squares) Plot shows that Root Mean PRESS is minimized when the number of factors is 7. This is stated in the note beneath the Root Mean PRESS Plot. A report called NIPALS Fit with 7 Factors is produced. A portion of that report is shown in Seven Extracted Factors.
The van der Voet T2 statistic tests to determine whether a model with a different number of factors differs significantly from the model with the minimum PRESS value. A common practice is to extract the smallest number of factors for which the van der Voet significance level exceeds 0.10 (SAS Institute Inc, 2011 and Tobias, 1995). If you were to apply this thinking here, you would fit a new model by entering 6 as the Number of Factors in the Model Launch panel.
|
8.
|
Select Diagnostics Plots from the NIPALS Fit with 7 Factors red triangle menu.
|
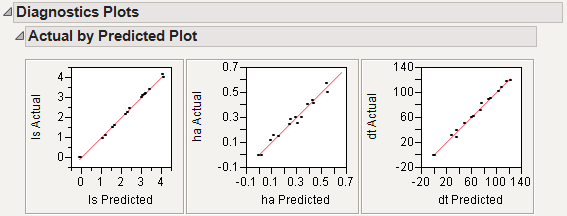
This gives a report showing actual by predicted plots and three reports showing various residual plots. The Actual by Predicted Plot in Diagnostics Plots shows the degree to which predicted compound amounts agree with actual amounts.
|
9.
|
Select VIP vs Coefficients Plot from the NIPALS Fit with 7 Factors red triangle menu.
|
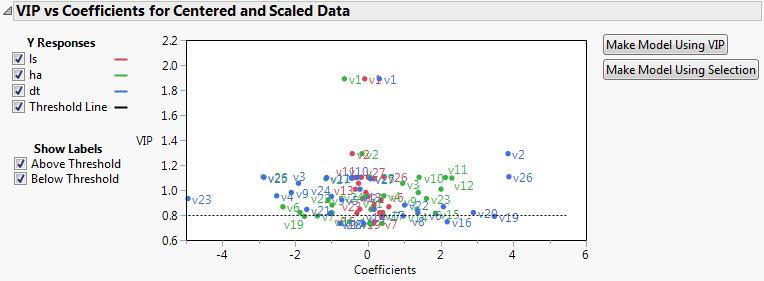
The VIP vs Coefficients plot helps identify variables that are influential relative to the fit for the various responses. For example, v23, v2, and v26 have both VIP values that exceed 0.8 and relatively large coefficients.
|
•
|
Select Analyze > Multivariate Methods > Partial Least Squares.
|
|
•
|
|
|
‒
|
In JMP Pro, you can enter nominal response columns in the Fit Model launch window to conduct PLS-DA. For details, see PLS Discriminant Analysis (PLS-DA).
Enter the predictor columns. The Partial Least Squares launch window only allows numeric predictors.
|
‒
|
Centers all Y variables and model effects by subtracting the mean from each column. See Centering and Scaling.
Scales all Y variables and model effects by dividing each column by its standard deviation. See Centering and Scaling.
(Fit Model launch window only) Select this option to center and scale all columns that are used in the construction of model effects. If this option is not selected, higher-order effects are constructed using the original data table columns. Then each higher-order effect is centered or scaled, based on the selected Centering and Scaling options. Note that Standardize X does not center or scale Y variables. See Standardize X.
Replaces missing data values in Ys or Xs with nonmissing values. Select the appropriate method from the Imputation Method list.
If Impute Missing Data is not selected, rows that are missing observations on any X variable are excluded from the analysis and no predictions are computed for these rows. Rows with no missing observations on X variables but with missing observations on Y variables are also excluded from the analysis, but predictions are computed.
(Appears only when Impute Missing Data is selected) Select from the following imputation methods:
|
‒
|
Mean: For each model effect or response column, replaces the missing value with the mean of the nonmissing values.
|
|
‒
|
EM: Uses an iterative Expectation-Maximization (EM) approach to impute missing values. On the first iteration, the specified model is fit to the data with missing values for an effect or response replaced by their means. Predicted values from the model for Y and the model for X are used to impute the missing values. For subsequent iterations, the missing values are replaced by their predicted values, given the conditional distribution using the current estimates.
|
After completing the launch window and clicking OK, the Model Launch control panel appears. See Model Launch Control Panel.
The Centering and Scaling options are selected by default. This means that predictors and responses are centered and scaled to have mean 0 and standard deviation 1. Centering the predictors and the responses places them on an equal footing relative to their variation. Without centering, both the variable’s mean and its variation around that mean are involved in constructing successive factors. To illustrate, suppose that Time and Temp are two of the predictors. Scaling them indicates that a change of one standard deviation in Time is approximately equivalent to a change of one standard deviation in Temp.
All model effects are then centered or scaled, in accordance with your selections of the Centering and Scaling options, prior to inclusion in the model.
If the Standardize X option is not selected, and Centering and Scaling are both selected, then the term that is entered into the model is calculated as follows: