The Cluster Subjects Across Study Sites report is used to identify similar subjects. It t does so by constructing a cross domain data set using as much data as possible (subject to user options). Next, it calculates Euclidean distances to compute a distance matrix and performs hierarchical clustering of subjects, across all of the study centers. Findings values are averaged by USUBJID, test code, visit number, and time point (if available) if there are multiple measurements for a visit or time point. The goal of this exercise is to identify pairs of subjects with a very small distance. This could be an indication that these subjects are in fact the same individual who has enrolled at multiple sites.
Running this report using the Nicardipine sample setting and default options generates the output shown below. Refer to the Cluster Subjects across Study Sites requirements description for more information. This report uses pre-dosing information with the goal of identifying subjects that have enrolled at two or more clinical sites.
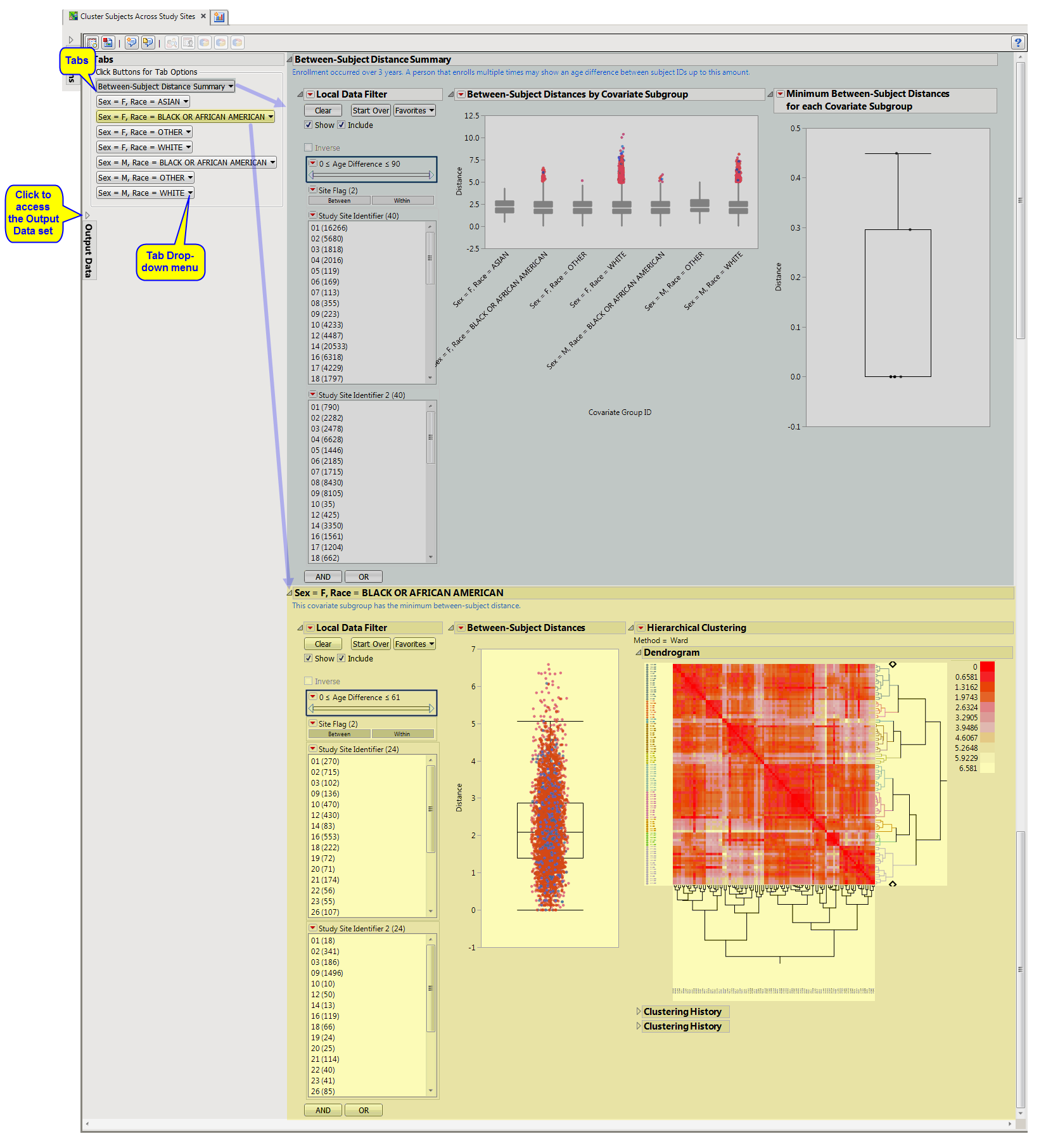
The Cluster Subjects Across Study Sites report shows the results of clustering of the subjects on the basis of different combinations of covariates (demographic groups in this example). The results for each grouping are presented on a separate “tab”.
This pane enables you to access and view the output plots and associated data sets on each tab. Use the drop-down menu to view the section in the Results pane or remove the section and its contents from the Results pane.
|
•
|
Between-Subject Distance Summary (Cluster Subjects Across Study Sites): Box plots are presented for all pairwise distances between subjects in the selected population. Pairs are limited based on selections from the Cluster subjects matching these criteria panel of the dialog.
|
In this example, box plots are presented by gender and race. The more similar a pair of subjects, the smaller the distance value (a zero indicates a perfect match). The minimum distance from each covariate subgroup is presented in the box plot to the right. The subgroup with the most similar pair of subjects is presented on the next tab.
|
•
|
One or more Subgroup Clustering sections: Only one tab is opened initially. The name of this tab is dependent on the covariate values used (as specified in the Cluster subjects matching these criteria panel) and the subgroup that is identified with the minimum pairwise distance.
|
In this example, the Sex =F, Race WHITE subgroup shows the minimum pairwise distance. Other subgroup results can be opened from the Results Sections sections menu.
|
•
|
Profile Subjects: Select subjects and click
|
|
•
|
Show Subjects: Select subjects and click
|
|
•
|
Show Rows in Heat Map: Select points that represent pairs of subjects in the Box Plots and click
|
|
•
|
Subset Clustering: On a subgroup clustering page, subsets clustering to subjects, based on pairs selected from corresponding box plot.
|
|
•
|
Revert Clustering: Click
|
|
•
|
Click
|
Output includes one summary data set (named csass_sum_XXX1, by default) containing one record per subject with pre-dosing data, one data set of all pairwise distances within the covariate subgroups (named csass_alldist_XXX, by default), one data set containing minimum pairwise distances for each covariate subgroup (named csass_mindist_XXX), by default), one data set per covariate subgroup containing pairwise distances (named csass_p_Y_XXX, by default, where Y is indexed 1 to the number of covariate subgroups) and one data set per covariate subgroup containing the distance matrix of subjects within the covariate subgroup (named csass_Y_XXX, by default, where Y is indexed 1 to the number of covariate subgroups).
|
•
|
Click
|
|
•
|
Click
|
|
•
|
Click
|
|
•
|
Click the arrow to reopen the completed report dialog used to generate this output.
|
|
•
|
Click the gray border to the left of the Options tab to open a dynamic report navigator that lists all of the reports in the review. Refer to Report Navigator for more information.
|
No testing is performed. Subjects are clustered within each site according to the selected clustering methodology. Refer to the JMP documentation on hierarchical clustering for statistical details.
The _XXX designation is used to designate a one- to three-digit number that is added sequentially to prevent overwriting of existing data sets.