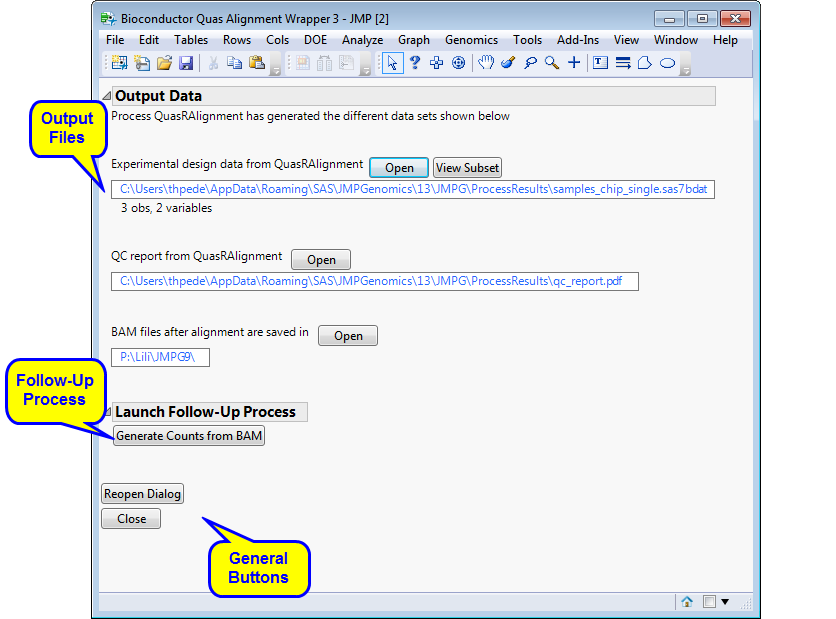
Running this process generates the Results window shown below. Refer to the Bioconductor QuasR Alignment Wrapper process description for more information.
The Results window contains the following elements:
|
•
|
Experimental Design Data Set (EDDS): This data set lists specific information about experimental details for each of the arrays. Refer to Experimental Design Data Set (EDDS) for more information about this data set. Click to view the data set.
|
|
•
|
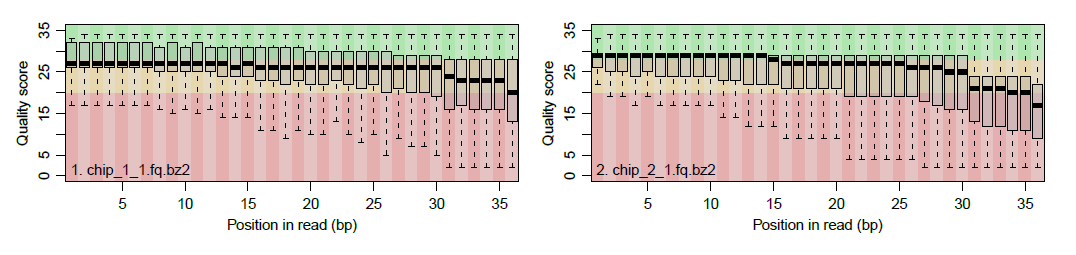
Quality Control report of the alignment: Shows plots on sequence qualities and alignment statistics. In the example shown below, the box plots (one for each chip) show the distributions of base quality values for each position in the input sequence. The background color (green, orange, or red) indicates ranges of high, intermediate, and low qualities. The plot is available for either fastq and BAM files.
|
|
•
|
A set of BAM files that can be imported for further analysis.
|
Note: You can view the data in a file or data set by clicking either or (when the data set is very large).
|
•
|
Generate Counts from BAM: Click to launch the BAM Input Engine process with the output BAM files specified as input.
|
|
•
|
Click to reopen the completed process dialog used to generate this output.
|
|
•
|
Click to close all graphics windows and underlying data sets associated with the output.
|