The Affymetrix Integrated Genome Browser process creates an .html table within JMP Genomics with embedded hyperlinks to chromosomal features and locations in the Affymetrix Integrated Genome Browser. Custom tracks can also be created for upload to the browser.
Before you can successfully use the Affymetrix Integrated Genome Browser process, you must first download the Integrated Genome Browser from the Affymetrix website (http://www.affymetrix.com/estore/partners_programs/programs/developer/tools/download_igb.affx). The browser requires that you have the JAVA Web Start application pre-installed. Several versions of the Genome Browser are available, different only in the amount of allocated memory required, from 256 MB to 1.5 GB.
Caution: Be careful not to choose a version that exceeds the amount of memory available for this process.
Refer to specific documentation, available on the same website, for information about downloading, installation and use of the Integrated Genome Browser. The browser must be open before you run the Affymetrix Integrated Genome Browser process.
|
•
|
chromosome and positional information for relevant genomic features, or
|
|
•
|
The data set can also contain one or more quantitative variables of interest that can be selected for inclusion in a custom track. such as:
|
•
|
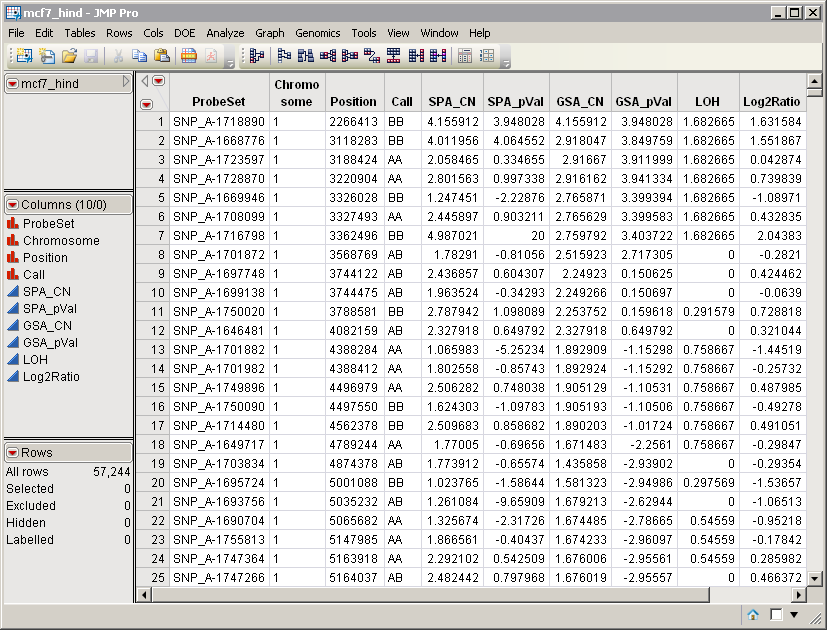
For an example, consider the mcf7_hind.sas7bdat data set.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
Refer to the Affymetrix Integrated Genome Browser output documentation for detailed descriptions of the output of this process.