The Illumina Methylation IDAT Input Engine imports a set of IDAT files and combines them into a single SAS data set. Before running this process, you must create an Experimental Design File (EDF) that indexes the IDAT files. This importer is based on R package illuminaio_0.8.0.
Note: This import process is considered experimental in this release of JMP Genomics.
|
•
|
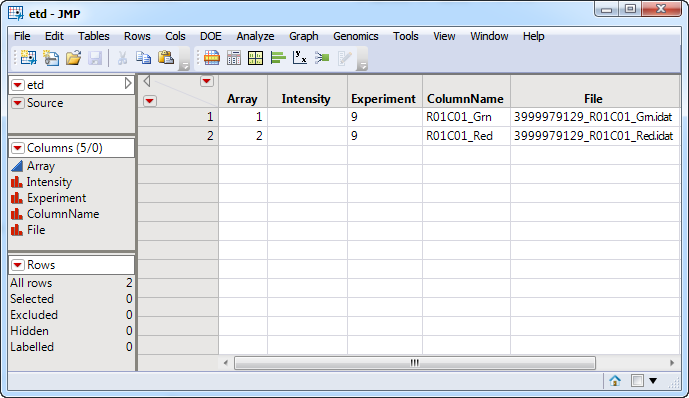
An Experimental Design File (EDF) that indexes the individual raw data files for the experiment. The EDF is typically a text file or Excel spread sheet and must be created before the data can be imported. A typical EDF, the etd.sas7bdat file, is shown below. This file specifies two input Illumina IDAT files.
|
|
•
|
Raw data files, which must be located and copied to a single folder. The raw data files must be binary files formatted in Illumina’s proprietary IDAT format with the .idat extension. There is typically a pair of input files, one file for each dye, for each sample.
|
The output data set includes mean values for each array. The output data sets generated by this process are listed in a Results window. Refer to the Illumina Methylation/Genotype IDAT Input Engine output documentation for detailed descriptions.