The Basic Genetics Workflow process builds and runs a basic workflow using a data set containing SNP genotype for a sample of unrelated individuals. This automated process uses the Marker Properties, Missing Genotype by Trait Summary, Subset and Reorder Genetic Data, Case-Control Association and the P-Value Quantile Plotter processes to perform the following analyses:
|
1
|
computations of various properties about the SNPs, such as minor allele frequency (MAF), proportion of missing genotypes at each SNP, and statistics for the test of Hardy-Weinberg equilibrium (HWE),
|
|
4
|
tests for association of individual SNPs with a binary trait using the Cochran-Armitage trend test, and
|
|
5
|
plotting of observed p-values from the trend test versus expected p-values under the null p-value distribution.
|
One wide-formatted SAS data set is required to run the Basic Genetics Workflow process. This data set must be formatted like those used in Affymetrix SNP CHP Input Engine or Illumina SNP Input Engine:
|
•
|
|
•
|
The samplegmdata.sas7bdat data set, described in Sample Genetic Marker Data, was computer generated and consists of 1000 rows of individuals with 130 columns corresponding to data on these individuals. This is a wide data set; molecular entities are listed in columns, and samples are listed in rows. Data for 60 markers is presented in the two-column allelic format in columns ma1-ma120. This data set also lists pedigree data, data for 4 different quantitative traits, and disease status.
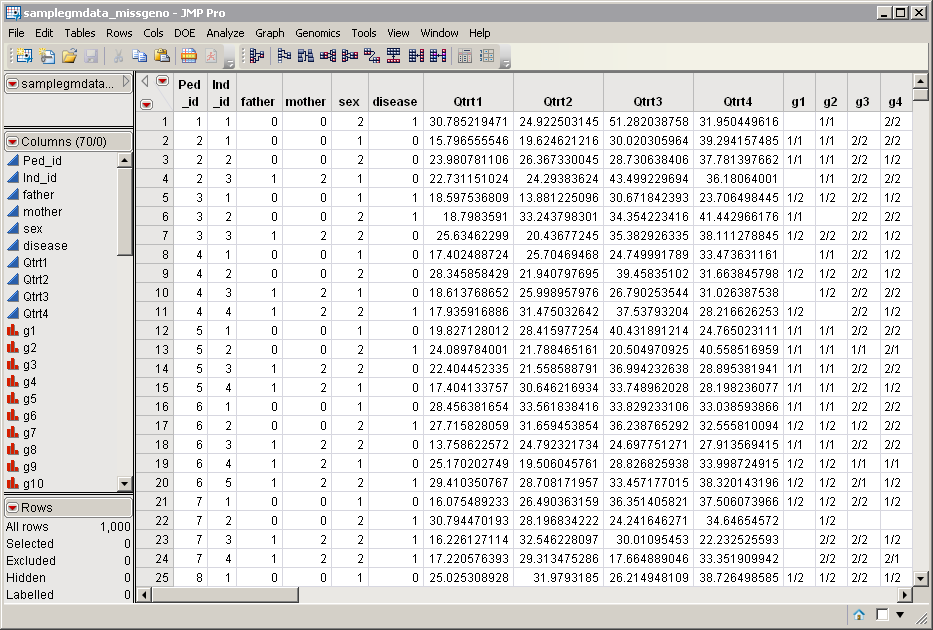
The samplegmdata_missgeno.sas7bdat data set is shown below. This data set was generated by making the following modifications to the samplegmdata.sas7bdat data set:
|
•
|
the marker data was converted from the two-column allelic format to the one-column genotypic format (60 columns, g1-g60), in which the alleles are delimited by a “/”, and
|
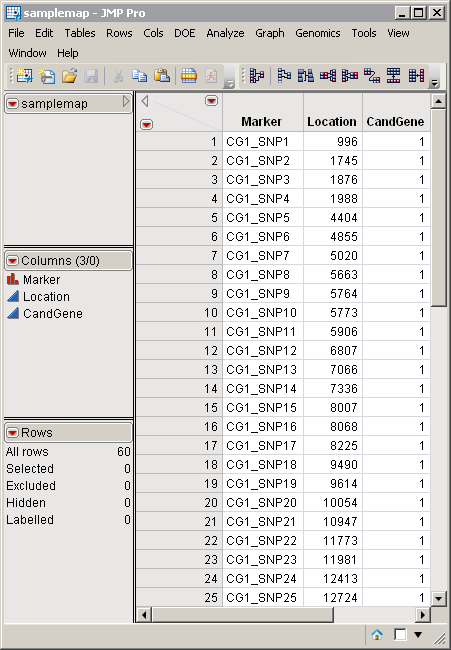
A second data set, the Annotation Data Set, is optional. This data set contains information such as gene identity or chromosomal location, for each of the markers. The samplemap.sas7bdat annotation data set serves as an example. It was computer generated and identifies markers, location, and gene identities. A portion of this data set is illustrated below. This data set is a tall data set; each row corresponds to a different marker.
Note: The top-to-bottom order of the rows in the annotation data set matches the left-to-right order of the columns in the input data set. This correspondence is required for markers to be matched appropriately.
Both the samplegmdata_missgeno.sas7bdat data set and the samplemap.sas7bdat annotation data set are located in the Sample Data\Genetics directory included with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
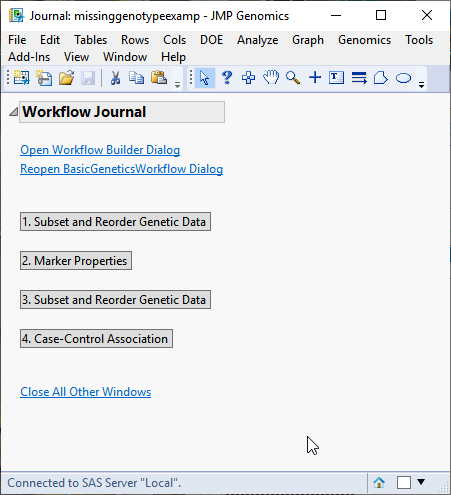
When you click , the Basic Genetics Workflow process begins by opening the Workflow Builder. The Workflow Builder builds a settings file for each process containing the information from the data sets and parameters specified in the Basic Genetics Workflow dialog. Once the setting files are generated and saved, the individual processes in the workflow are sequentially opened, populated, and run. The results of the processes are saved in the specified output folder. Finally, a JMP journal, providing links to the workflow dialog and the results of each process, is generated. The Journal is shown below.
|
|
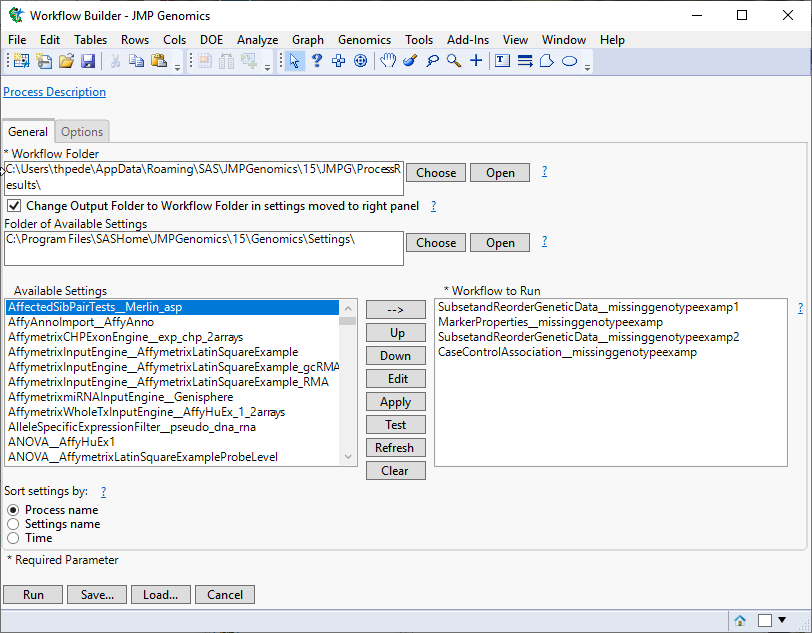
Click to view the Workflow Builder.
|
The Workflow Builder dialog shows the settings for each of the processes in the workflow. You can select and edit individual settings to adjust your analysis.
|
|
Click under any of the processes shown in the journal to view the output for those processes. For your convenience, links to default Basic Genetics Workflow processes are given below.
|