The Genetics Rare Variants Workflow process runs a workflow for performing association testing on both common and rare variants, automatically coding rare variants according to a dominant model, and optionally combining rare variants within a gene or other defined set of SNPs into a single locus. Association tests can then be performed on each single locus, or combined tests can be performed on all common and rare variants within a gene or SNP set. Both the Combined Multivariate and Collapsing (CMC) method described by Li and Leal (2008) and the Rare Variant Test 2 (RVT2) described by Morris and Zeggini (2009) can be implemented using this workflow. For binary or continuous traits, the Sequence Kernel Association Test (SKAT) is also an option.
|
1
|
|
2
|
and several processes for performing association testing of SNPs with the trait variables specified:
|
3
|
Case-Control Association, to perform Pearson chi-square and Fisher's exact test on the numeric genotypes,
|
|
5
|
One wide-formatted input SAS data set that contains all of the marker data is required to run the Genetic Rare Variants Workflow process. This data set must be formatted like those using the Affymetrix SNP CHP Input Engine or Illumina SNP Input Engine: marker variables contain “/”-delimited genotypes (such as A/A, A/B).
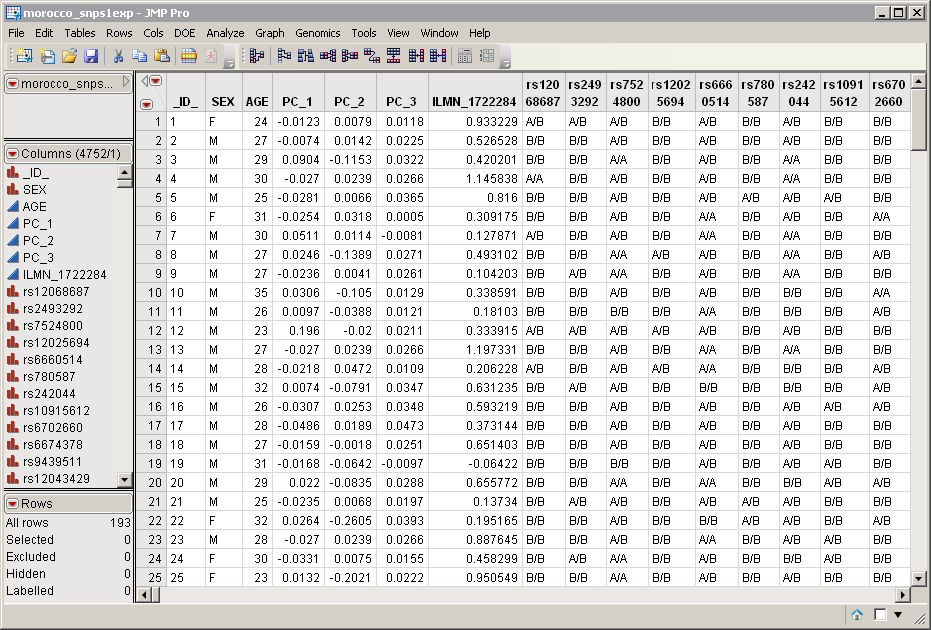
The morocco_snps1exp.sas7bdat data set represents the data from a study of gene expression variation and SNP associations in southern Morocco (Idaghdour, Czika, et al., 2010), and serves as an example. (Note: The data was modified slightly to preserve anonymity of subjects.) It lists genotype data at 4746 SNPs in 193 individuals. Marker data is presented in the one-column format. This data set is partially shown below. Note that this is a wide data set; markers are listed in columns, whereas individuals are listed in rows.
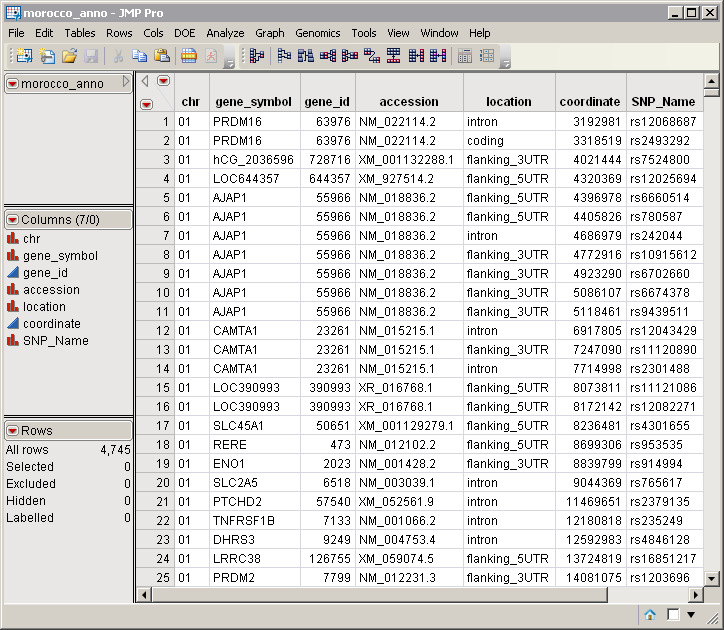
A second, optional, data set is the Annotation Data Set. This data set contains information such as gene identity or chromosomal location, for each of the markers. The morocco_anno.sas7bdat annotation data set serves as an example. It identifies markers, location, and gene identities. A portion of this data set is illustrated below. This data set is a tall data set; each row corresponds to a different marker.
Note: The top-to-bottom order of the rows in the annotation data set matches the left-to-right order of the columns in the input data set. This correspondence is required for markers to be matched appropriately.
Both the morocco_snps1exp.sas7bdat data set and the morocco_anno.sas7bdat annotation data set are located in the Sample Data\Genetics directory included with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
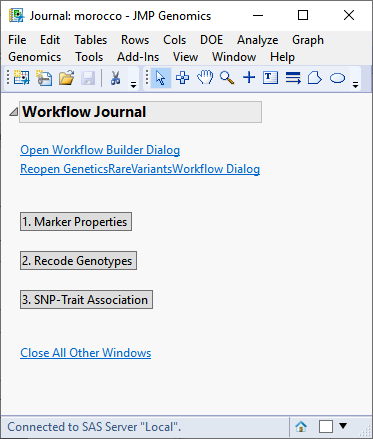
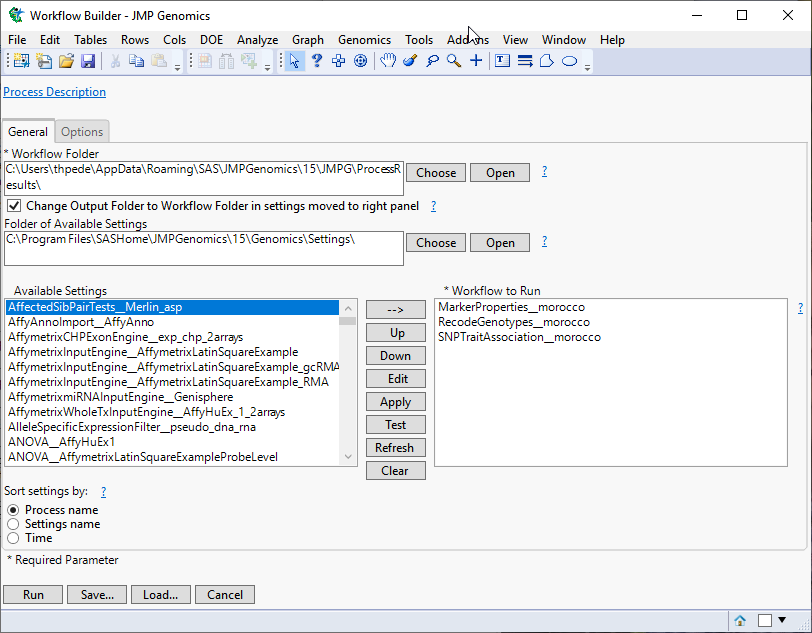
When you click , the Genetics Rare Variants Workflow process begins by opening the Workflow Builder. The Workflow Builder builds a settings file for each process, containing the information from the data sets and parameters specified in the Genetics Rare Variants Workflow dialog. Once the setting files are generated and saved, the individual processes in the workflow are sequentially opened, populated, and run. The results of the processes are saved in the specified output folder. Finally, a JMP journal, providing links to the workflow dialog and the results of each process, is generated.
|
|
Click .
|
The Workflow Builder dialog shows the settings for each of the processes in the workflow. You can select and edit individual settings to adjust your analysis.
|
|
Clicking each of the buttons on the journal brings up the output of each of the processes. This enables you to examine each set of output so that adjustments can be made to the individual settings, as needed. For your convenience, links to the default Genetics Rare Variants Workflow processes are given below.
|