The Basic Linkage Mapping Workflow process builds and runs a workflow to create a genetic linkage map using marker genotypes for individuals or lines from an experimental inbred population. This automated process runs the Recombination and Linkage Groups, Linkage Map Order, Linkage Map Viewer, and Subset and Reorder Genetic Data processes to perform the following analyses:
|
1
|
Compute recombination frequencies between all pairs of markers, test for marker segregation distortion, and assign markers into linkage groups using automated Hierarchical Clustering (this algorithm is used by default if a framework map has not been specified) or Minimum Recombination Grouping (this algorithm is used when a framework map is used to accommodate the constraints of markers in known groups).
|
|
2
|
Run the Map Order optimization algorithm to determine the optimal order of the markers within the generated linkage groups and compute the genetic distance map based on the pairwise recombination frequencies.
|
|
4
|
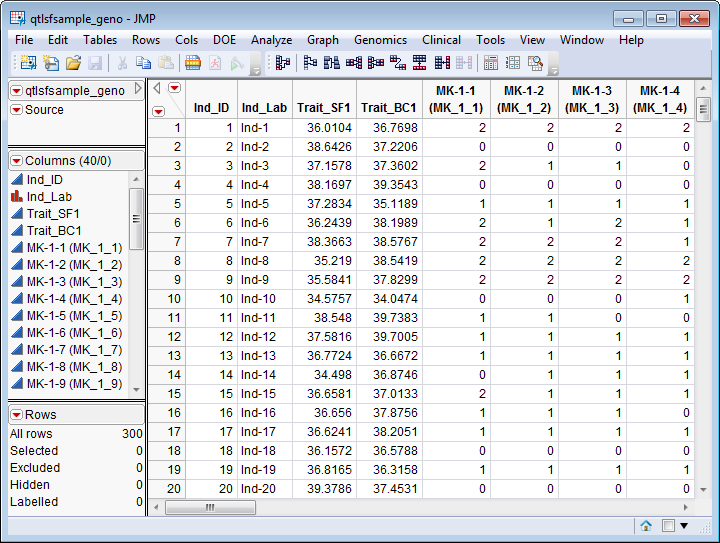
An Input Genotype SAS Data Set is required for this process. The qtlsfsample_geno.sas7bdat data set serves as an example, and is shown below. It contains 300 rows of individuals and 40 columns corresponding to identifier, trait, and marker genotype information.
Optional data sets include:
|
•
|
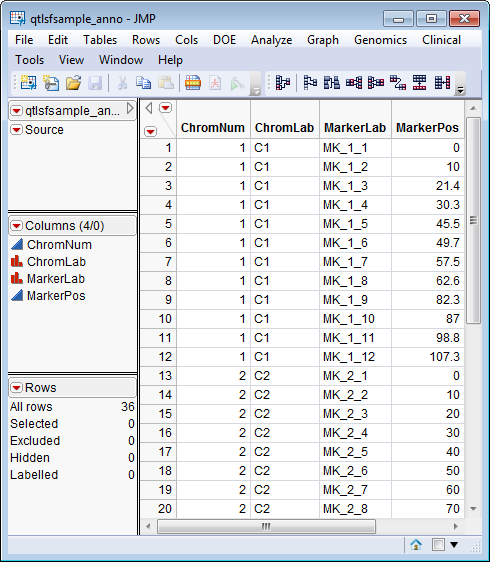
An Annotation SAS Data Set. This data set contains information such as gene identity or chromosomal location, for each of the markers. The qtlsfsample_anno.sas7bdat data set serves as an example, and is shown below. It contains 36 rows of markers and 4 columns containing chromosome, marker, and positioning information.
|
|
•
|
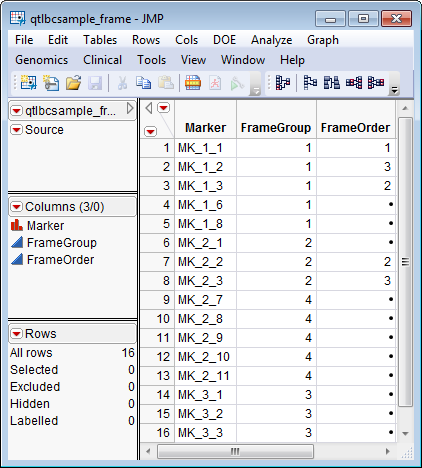
A Framework Map Data Set. This data set can be used to create linkage groups for markers in the genotype data that incorporate prior knowledge of markers that already belong to pre-defined groups. The qtlbcsample_frame.sas7bdat data set serves as an example, and is shown below.
|
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
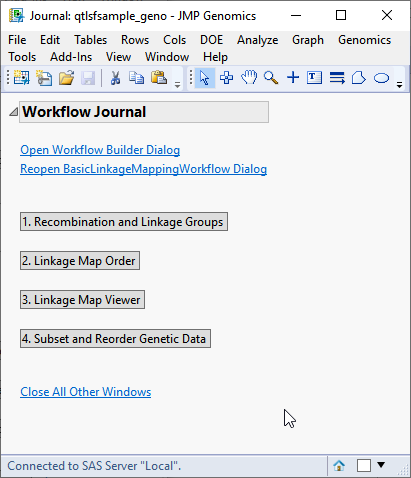
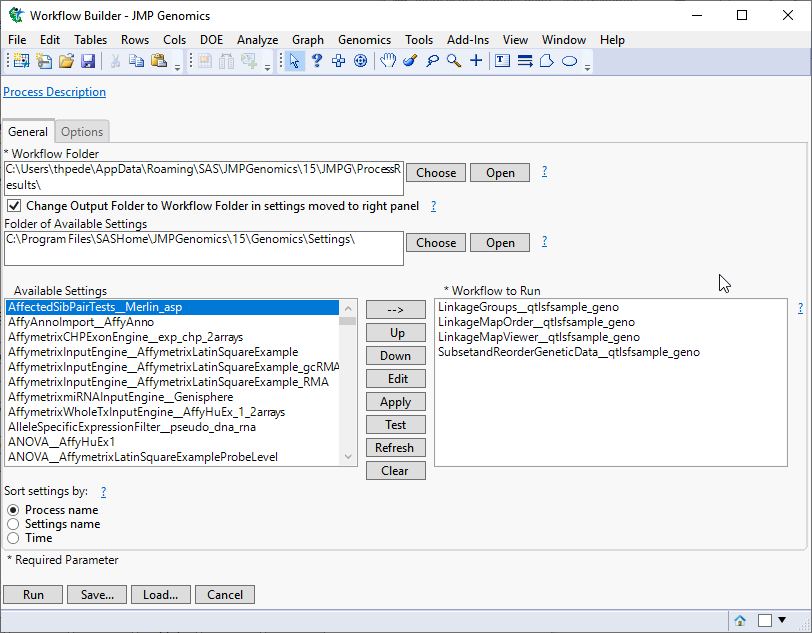
When you click , the Basic Linkage Mapping Workflow process begins by opening the Workflow Builder. The Workflow Builder builds a settings file for each process, containing the information from the data sets and parameters specified in the Basic Linkage Mapping Workflow dialog. Once the setting files are generated and saved, the individual processes in the workflow are sequentially opened, populated, and run. The results of the processes are saved in the specified output folder. Finally, a JMP journal, providing links to the workflow dialog and the results of each process, is generated.
|
|
Click .
|
The Workflow Builder dialog shows the settings for each of the processes in the workflow. You can select and edit individual settings to adjust your analysis by highlighting the desired workflow and selecting .
|
|
Clicking each of the buttons on the journal brings up the output for each of the processes. This enables you to examine each set of output so that adjustments can be made to the individual settings, as needed. For your convenience, links to default Basic Linkage Mapping Workflow processes are given below.
|