Output Description
Affymetrix Exon CHP Input Engine
Running this process for the Affymetrix Human Exon 1.0 ST colon cancer data set generates the Results window shown below. Refer to the Affymetrix Exon CHP Input Engine process description for more information.
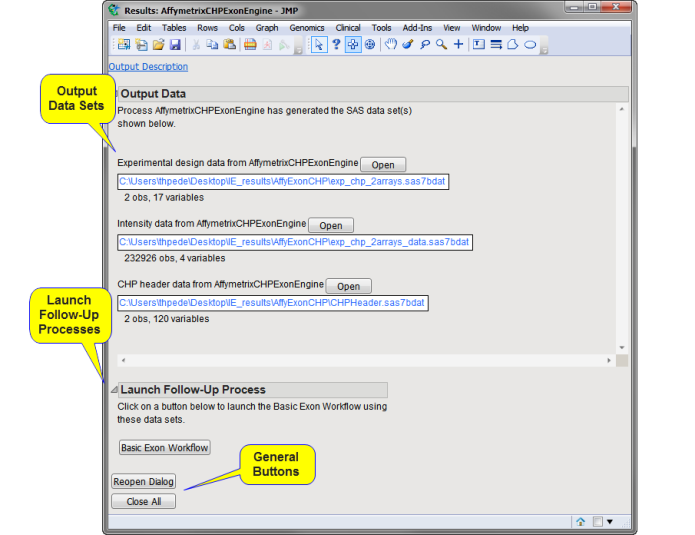
The Results window contains the following elements:
Output Data
This process generates the following data sets:
| • | Experimental Design Data Set (EDDS): This data set lists specific information about experimental details for each of the arrays. Refer to Experimental Design Data Set (EDDS) for more information about this data set. |
| • | Output Data Set: This data set contains the data from the different .chp files. This data set is in the tall format: the intensity data from each array are listed in a separate column while individual spots are listed in rows. |
| • | CHP Header Data Set: This data set contains all the parameters in the header portion of each Command Console file. These parameters include the algorithm applied, the type of chip used, the program applied, and other quality control information among the control l probes. The parameters are listed in columns, while the individual .cel files are listed in rows. |
Note: You can view the data in a file or data set by clicking either or (when the data set is very large).
Tip: To filter the Quality Flag data set directly using the Detection_Percentage column, use the Filter Intensities process. Replace all values in the Detection_Percentage column falling below an acceptable percentage with the missing value symbol (.) and then delete all rows with a missing value symbol. Merge the filtered QualityFlag data set back to the main data table, using the Merge process.
Launch Follow-up Processes
| • | Basic Exon Workflow: Click to launch the Basic Exon/Alternative Splicing Workflow process with the output data set and the EDDS specified as input. |
General
| • | Click to reopen the completed process dialog used to generate this output. |
| • | Click to close all graphics windows and underlying data sets associated with the output. |