Results
This tab provides a comprehensive summary of ANOVA model fitting results. It is important to keep in mind which model was fit and to carefully consider hypotheses of interest. Depending on the variability in your data and your objectives, you might wish to alter the significance criterion to obtain fewer or more significant genes. The numerous drill-down options available in the Action Buttons pane on the left-hand side are valuable for exploring interesting subsets.
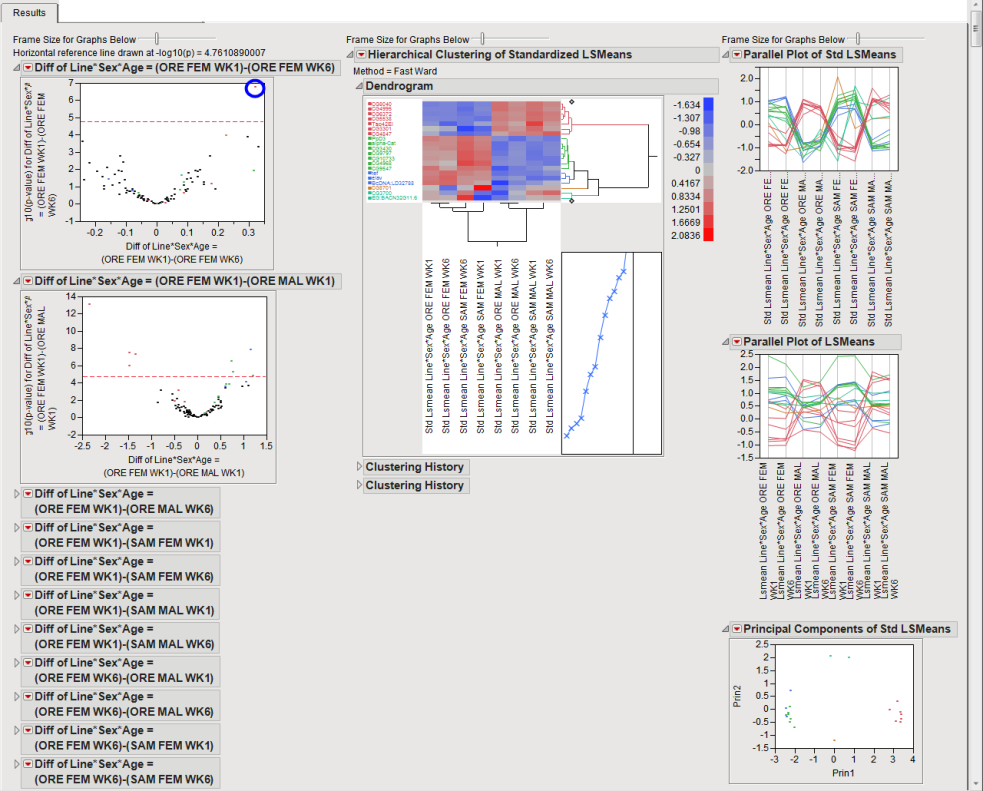
The Results tab contains the following elements:
| • | A series of Volcano Plots. |
Volcano plots are a convenient way to summarize a specific hypothesis test across all genes. Each plot is based on a single hypothesis of interest and each point in the plot is a gene. The x-axis represents a difference or estimate and the y-axis its corresponding -log10(p-value). Volcano plots have a characteristic "V" shape because estimates near zero tend not to be significant and those away from zero tend to have smaller p-values / larger -log10(p-values). Significant genes are those in the upper left and right quadrants of the plot, akin to exploding pieces of molten lava. The red dashed horizontal line usually represents a significant criterion computed by some multiple testing method like FDR. You can change this value with an action button in the left panel. You can also resize all of the plots with a slider above them.
When interpreting volcano plots, it is important keep in mind the direction of the difference on the x-axis. For example, the first plot from the Drosophila aging example has Diff of Line*Sex*Age = (ORE FEM WK1) - (ORE FEM WK6) on the x-axis. One gene is significant in the upper right corner of the plot (circled). Considering the direction of the difference, this indicates that this gene is significantly down-regulated in week 6 as compared to week 1 for Oregon females.
You can mouse over points of interest to see their labels or select points by dragging a mouse rectangle over them. Use the lasso tool to select irregular regions. To find specific genes whose identifier you know, click Results in the Tabs section then View Data. In the subsequently opened data table, click Edit > Search, and type in the desired search string. Any genes that you select in the table are highlighted in the graphs and vice versa. Selected genes are highlighted in other plots and you can also then click on various Action Buttons on the left-hand side for further analyses on those specific genes.
Volcano plots are generated for the set of LS means you specify in the input dialog (for example, all possible pairs or differences with a control) as well as for all custom ESTIMATE statements that you specify.
See Volcano Plot for more information.
| • | A dendrogram showing the Hierarchical Clustering of Standardized LSMeans |
This plot enables you to compare expression patterns for all significant genes simultaneously. The standardized least squares means for every gene that is significant in at least one volcano plot are clustered both horizontally and vertically and depicted with a heat map. The standardization is to mean 0 and variance 1, so the emphasis is on patterns and not overall expression. Each row of the heat map is a gene and each column is a distinct LS mean. You can see which genes and LS means have similar expression profiles. You can click on branches of the horizontal dendrogram to select all genes in that cluster. These genes are then highlighted in other plots and you can then click Action Buttons on the left-hand side for further analyses.
Click and slide the cross-hair point at the top or bottom of the horizontal dendrogram to change the number of colored cluster groups.
See Heat Map and Dendrogram for more information.
| • | A parallel plot of Standardized LSMeans. |
This plot shows the same data as the hierarchical clustering plot, but in the form of a parallel plot. Note the x-axis is in a sorted order and is usually different from the order in the hierarchical clustering plot. You can mouse over lines of interest to select genes.
See Parallel Plot for more information.
| • | A parallel plot of LSMeans. |
This plot shows the unstandardized LS means, enabling you to see degree of overall expression for the same set of significant genes.
See Parallel Plot for more information.
| • | Principal Components of Standardized LSMeans |
This plot provides an alternative way of comparing significant standardized LS means. It computes a principal components analysis on them and plots the first two components. This projects high-dimensional patterns into two dimensions. Genes that cluster together in this plot tend to also cluster together in the hierarchical clustering and parallel plots. This plot can help identify outliers. Points near the outer virtual bounding ellipse are well-explained by the first two principal components.
See Principal Components Analysis Plot for more information.
This tab provides a comprehensive summary of ANOVA model fitting results. It is important to keep in mind which model was fit and to carefully consider hypotheses of interest. Depending on the variability in your data and your objectives, you might wish to alter the significance criterion to obtain fewer or more significant genes. The numerous drill-down options available in the Action Buttons pane on the left-hand side are valuable for exploring interesting subsets.