Annotation Group Results (Missing Genotypes by Trait Summary)
The Annotation Group Results tab is shown below:
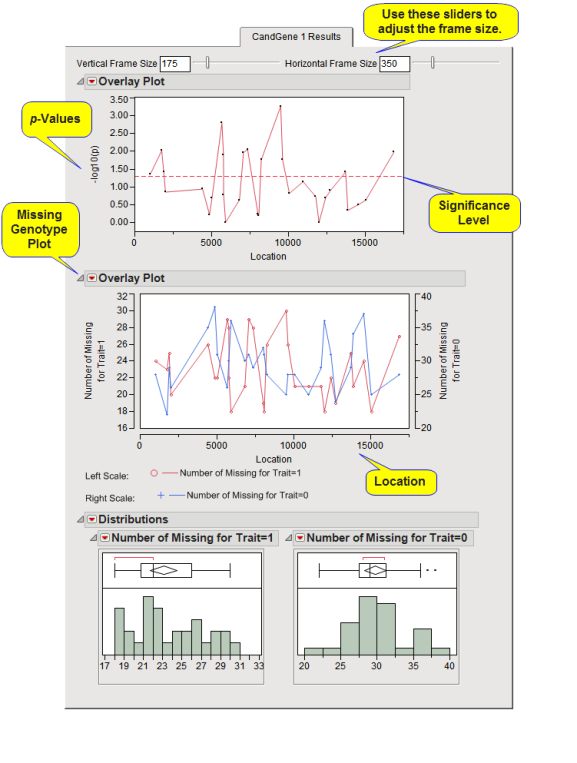
The Annotation Group Results tab contains the following elements:
| • | One or more P-Value Plots. |
The y-axis variable is the p-value from the missing genotype by trait test, converted to the -log or -log10 scale if selected in the process dialog. The x-axis plots location of the marker according to the annotation location variable within the annotation group.
A horizontal reference line is drawn as a red, dashed line at the significance level that was specified. For -log or -log10-converted p-values, markers above this line are significant; for p-values on the original scale, markers below the line are significant.
If any BY variables were selected, a separate chart is displayed for each BY group.
| • | One or more count of missing genotype overlay plot. |
For each value of the Trait Variable, the number of missing genotypes is plotted on the y-axis, and the x-axis plots location of the marker according to the annotation location variable within the annotation group (like the plot above).
If any By Variables were selected, a separate chart is displayed for each BY group.
| • | One or more count of missing genotype distribution charts. |
For each value of the Trait Variable, the number of missing genotypes is displayed in a frequency chart. If any By Variables were selected, a separate set of charts is displayed for each BY group.
The action buttons can be used on markers selected from these tabs as described for the Manhattan Plots.