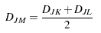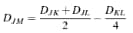Optimized Automated Clustering Method
Use the drop-down menu to specify the method to use for hierarchical clustering via PROC CLUSTER.
Note: This parameter is available only when either Optimized or Automated has been selected as the Compression Method.

Clustering methods are described in the following table:
|
Clustering Method |
Description |
||||||
|
Average |
Choose this method to set the distance between clusters to the average distance between pairs of observations. This method tends to join clusters with small variances and is biased toward producing clusters with the same variance.1 |
||||||
|
Centroid |
Choose this method to set the distance between clusters to the squared Euclidean distance between the mean of each cluster.2 This method is more robust than other clustering methods. |
||||||
|
Complete |
Choose this method to set the distance between clusters to the maximum distance between an observation in one cluster and an observation in the other.2 This method is biased toward producing clusters of equivalent diameters and can be distorted by even moderate outliers. |
||||||
|
Density |
Choose this method to use nonparametric probability density estimates (for example, Hartigan, 19753 (pp. 205–212); Wong, 19824; Wong and Lane 19835). Density linkage consists of two steps:
The CLUSTER procedure supports three types of density linkage: the kth-nearest-neighbor method, the uniform-kernel method, and Wong’s hybrid method. |
||||||
|
Flexible |
Choose this method to use the following combinatorial formula for clustering:
The flexible-beta method was developed by Lance and Williams (1967)6. |
||||||
|
McQuitty |
Choose this method to use the following combinatorial formula for clustering:
The method was independently developed by Sokal and Michener (1958)7 and McQuitty (1966)8. |
||||||
|
Median |
Choose this method to use the following combinatorial formula for clustering:
The median method was developed by Gower (1967)9. |
||||||
|
Single |
Choose this method to set the distance between two clusters to the minimum distance between an observation in one cluster and an observation in the other cluster. Because there are no constraints on the shape of clusters, single linkage sacrifices performance in the recovery of compact clusters in return for the ability to detect elongated and irregular clusters. Single linkage tends to chop off the tails of distributions before separating the main clusters. |
||||||
|
Twostage |
This option is a modification of density linkage that ensures that all points are assigned to modal clusters before the modal clusters are permitted to join. The CLUSTER procedure supports the same three varieties of two-stage density linkage as of ordinary density linkage: kth-nearest neighbor, uniform kernel, and hybrid. In the first stage, disjoint modal clusters are formed. The algorithm is the same as the single linkage algorithm ordinarily used with density linkage. In the second stage, the modal clusters are hierarchically joined by single linkage. The final number of clusters can exceed one when there are wide gaps between the clusters or when the smoothing parameter is small. |
||||||
|
Ward |
Choose this method to set the distance between clusters to the ANOVA sum of squares between the two clusters summed over all the variables. At each generation, two clusters from the previous generation are merged to reduce the within-cluster sum of squares over all partitions. The sums of squares are easier to interpret when they are divided by the total sum of squares to give the proportions of variance (squared semipartial correlations). This method joins clusters to maximize the likelihood at each level of the hierarchy under the assumptions of multivariate normal mixtures, spherical covariance matrices, and equal sampling probabilities. This method tends to join clusters with a small number of observations and is biased toward producing clusters with approximately the same number of observations. It is also very sensitive to outliers.2 |
Your choice of method might require additional options to be specified in the Additional PROC CLUSTER Options text field on the Options tab. The following is a brief list of methods that require or recommend additional parameter specifications: COMPLETE (TRIM= recommended); DENSITY (K=,R=, or HYBRID option must be specified); FLEXIBLE (See BETA= Option); TWOSTAGE (K=,R=, or HYBRID option must be specified); and WARD (TRIM= recommended) .
To Specify a Clustering Method:
| 8 | Specify Optimized as the Compression Method. |
| 8 | Make a selection using the drop-down menu. |
You should refer to the SAS PROC CLUSTER documentation for details about all of these methods.