Process Description
Pseudo Image
Genomic data sets often contain values or observations that deviate substantially from expected values. These deviations can often be ascribed to a variety of technical or other factors that have little to do with biological cause and effects. In these situations, it is often better to identify and exclude outliers from an analysis than it is to include them, particularly where the cause of the anomalies can be explained by technical problems.
Often, valuable information about the quality of an array can be obtained from a visual examination of an image of the hybridization pattern. The Pseudo Image process generates an interactive pseudo image of an array by plotting a color-coded intensity variable using X- and Y-coordinates.
What do I need?
Three data sets are required for this process.
The first required data set is the Experimental Design Data Set (EDDS). The affylatin_exp2.sas7bdat EDDS, which is used in the example that follows, is shown below. This required data set provides information about the experiment and identifies the files and columns containing the relevant data in the input data set. Note that one column in the EDDS must be named ColumnName and the values contained in this column must exactly match the column names in the input data set.

The second required data set, the Input Data Set, contains all of the numeric data to be analyzed. The sample data set used in the following example, the chips45and55.sas7bdat data set, shown below, is from the Affymetrix Latin Square experiment that is described in Sample Case Studies. This data was originally generated by Affymetrix Corporation to develop and validate their U95A GeneChip and Microarray Suite (MAS) 5.0 algorithm over a range of known concentrations. Each group in the experiment contains a pool of non-specific RNA as well as a set of 14 distinct human transcripts spiked in at known concentrations. Intensity data are listed in the last two columns of the data set.

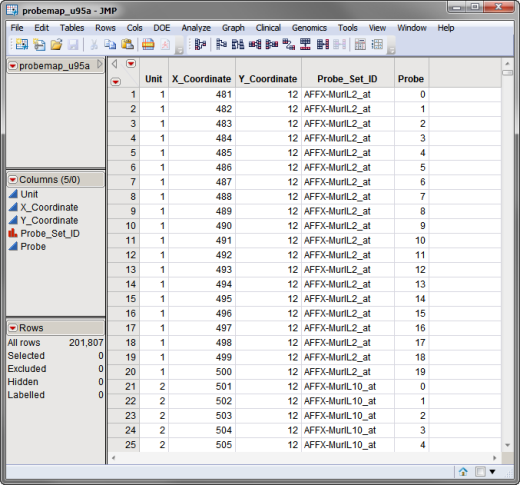
The third required data set, the coordinate data set, lists the x- and y-coordinates for each spot on the arrays. The probemap_u95a.sas7bdat data set, used in the following example, is shown below.
Two additional, optional, data sets can also be merged into the analysis. The first, the Annotation Data Set, provides annotation information for each of the markers, such as gene names, function, physical location, and association, that can be used as key terms for web and database searches.
The second optional data set, the QC data set, contains Affymetrix Input Engine generated QC probes data (qcprobe). This data set should be specified only if the input data set does not contain data for QC probes and if you want QC probes data displayed in the pseudo image. The qcdata_c45and55.sas7bdat QC data set is shown below.

| 8 | Examine and compare the chips45and55.sas7bdat input data set, the probemap_u95a.sas7bdat coordinate data set, and the qcdata_c45and55.sas7bdat QC data set. |
All three data sets have the Unit, Probe, and Probe_Set_ID columns in common.
The affylatin_exp2.sas7bdat EDDS, the chips45and55.sas7bdat input data set, the probemap_u95a.sas7bdat coordinate data set and the qcdata_c45and55.sas7bdat QC data set are all included in the Sample Data folder.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the Pseudo Image output documentation for detailed descriptions and guides to interpreting your results.