Process Description
Difference Chooser
The Difference Chooser process provides you with a quick and relatively easy method for selecting LSMeans fixed effect level differences for inclusion in both ANOVA and mixed model analyses. Differences are obtained from a SAS PROC Mixed LSMEANS statement and built from the Experimental Design Data Set (EDDS). These differences are then saved to a new SAS data set, which can be used as input to both anova and mixed model analyses, as well as those Basic workflows that call the ANOVA process.
What do I need?
One data set, an Experimental Design Data Set (EDDS), is needed to run this process. This required data set tells how the experiment was performed, providing information about the columns of the primary experimental data. The drosophilaaging_exp.sas7bdat EDDS, associated with the Drosophila aging experiment from (Jin, et al., 2001) and described in Sample Case Studies, is used in the example that follows. This data set is shown below.
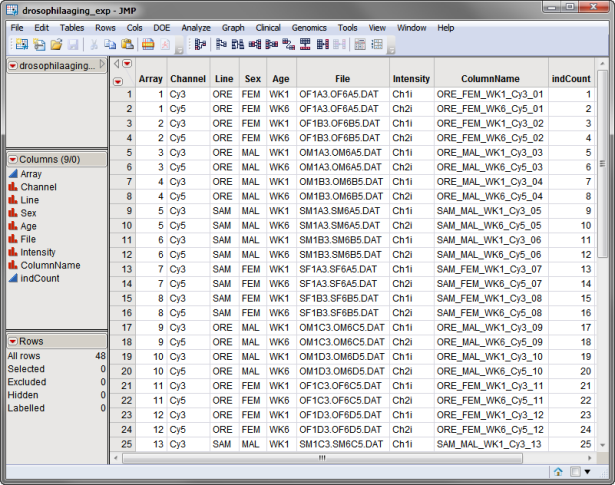
This data set is located in the Sample Data folder.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Example
This example uses the drosophilaaging_exp.sas7bdat EDDS shown above.
| 8 | Click to select the experimental design data set (EDDS). |
| 8 | Navigate into the SampleData \Microarray\Scanalyze Drosophila\ directory. |
| 8 | Select the drosophilaaging_exp.sas7bdat file and click . |
The drosophilaaging_norm.sas7bdat file has been selected. If you click , the data set illustrated above opens.
All of the variables in the data set are displayed in the Available Variables field. In this experiment, Line, Sex, Age, Array, and Channel all function as class variables.
| 8 | Examine the list of available variables. Highlight Line, Sex, Age, Array, and Channel. |
| 8 | Click  to add Line, Sex, Age, Array, and Channel to the Class Variables field. to add Line, Sex, Age, Array, and Channel to the Class Variables field. |
The Difference Chooser focuses on the effects that different LSMeans effects have, both individually and in combination, on gene expression. To specify potentially interacting factors, complete the following steps:
| 8 | Type line*sex*age to allow for the investigation of the interaction of these three factors. |
| 8 | Make sure that All Pairwise Differences is selected as the LSMeans difference set to be generated. |
Because Differences with a Control was not selected as the LSMeans Difference Set, the LSMeans Control Levels field is inactive.
| 8 | Leave the Output Data Set field blank. |
To specify an output folder, complete the following steps:
| 8 | Click . |
| 8 | Navigate to the folder where you want your results placed. |
| 8 | Click . |
The completed dialog should appear as shown below.

| 8 | Click to generate a second dialog window. |
The Difference Chooser GUI window appears, as shown below.

Examine the Difference Chooser GUI window. Note that all of the pairwise comparisons are listed in the window. This window is used to specify which set of LSMeans differences are to be tested using anova or mixed model analysis. You can also reverse the order of the comparisons.
| 8 | Check the boxes to Include the first two differences, as shown below. |
You must save the LSMeans differences to the output data set created by the Difference Chooser.
| 8 | Click to save the differences. |
The name and path to the output data set are illustrated in a JMP Genomics Message window, as shown below.

Output/Results
The output of this process is a file containing the LSMeans fixed-effect-level differences. This file can be used in subsequent analyses (for example, ANOVA or Mixed Model Analysis) of the experimental data.
