Process Description
TPM Normalization
The TPM Normalization process (Transcripts Per Kilobase Million) is a normalization method for Count data that takes reads per kilobase (RPK) and adjusts per million scaling factor for each sample to generate the TPM.
One benefit of TPM normalization is that the sum of all TPMs in a sample are equivalent, making comparisons of the number of reads for a gene across samples easier.
What do I need?
Two data sets are required to run this process.
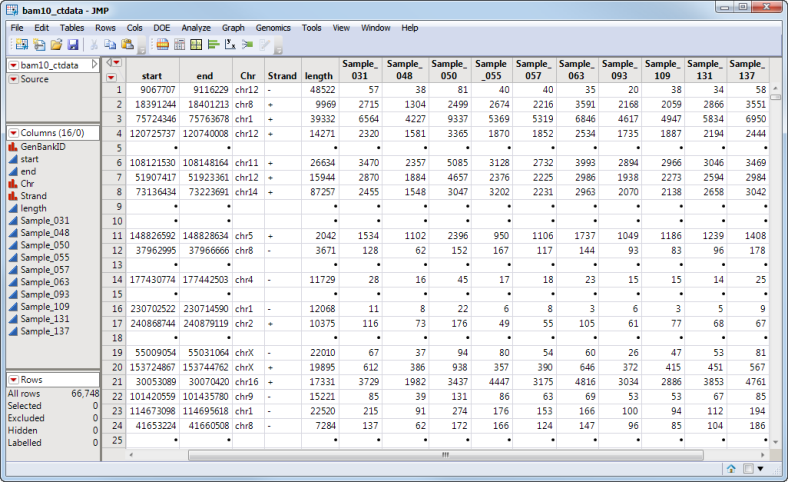
The first data set, the Input Data Set, contains all of the numeric data to be analyzed. This data set must be in the tall format where each sample corresponds to one row and each column corresponds to a separate experimental condition or array.
The trimmed bam10_ctdata.sas7bdat data set shown below lists sample BAM data.
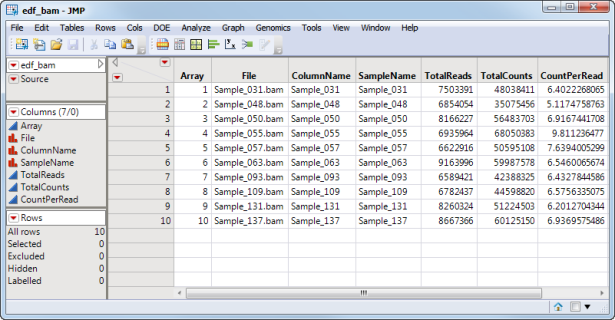
The second data set is the Experimental Design Data Set (EDDS). This required data set tells how the experiment was performed, providing information about the columns in the input data set. Note that one column in the EDDS must be named ColumnName and the values contained in this column must exactly match the column names in the input data set.
The edf_bam_sas7bdat EDDS, shown below, corresponds to the bam10_ctdata.sas7bdat input data set.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
Refer to the TPM Normalization output documentation for detailed descriptions of the output of this process.