Process Description
Affected Sib-Pair Tests
The Affected Sib-Pair Tests process enables you to perform various affected sib-pair chi-square tests of linkage for a dichotomous trait. See Elston and Cordell (2001, pp.142-143) and Whittemore and Tu (1998) for a summary of these tests.
The Affected Sib-Pair Tests process is not applied to genetic marker data like the other JMP Genetic processes. Instead, this process analyzes data containing information about the probabilities of pairs of individuals sharing alleles that are identical-by-descent (IBD) at the markers of interest. The required input IBD data set must contain one row for each pair of related individuals being analyzed at each marker, with variables z0, z1, and z2 representing the probability of the two individuals in the pair sharing 0, 1, or 2 alleles IBD, respectively. All possible pairwise comparisons within each family should be made. Variables for the pedigree or family, the two individual IDs, and the marker are also required in this data set. Pairs of individuals should be grouped by marker, then by pedigree or family before carrying out these processes.
What do I need?
One data set, the identical by descent (IBD) data set, which contains the marker data, pedigree ID, and the IBD probabilities is required for this process. This data set must contain the following variables: Marker, a pedigree ID, and the IBD probabilities in columns z0, z1, and z2.
The sample data set used in the following example, comes from the computer-generated affected sib-pair (ASP) data provided by Gonçalo Abecasis (University of Michigan Center for Statistical Genetics) and described in Affected Sib-Pair (ASP) Data. This data includes three associated data sets:
1 The IBD data set contains the IBD probabilities for 20 markers in 200 families, with 4 individuals in each family: two unrelated parents (individuals 1 and 2) and two full-sib progeny (individuals 3 and 4) of individuals 1 and 2.
| 2 | A pedigree data set that lists the family relationships, affected status, and marker genotype for each of the 800 individuals (4 per family) in the data set, and |
| 3 | A map data set that lists the physical location of each of the markers on human chromosome 24. |
Note: If you are curious about chromosome 24, recall that these are fictitious data.
The asp_ibd.sas7bdat IBD data set is shown below. Note that the required columns are present in this data set.

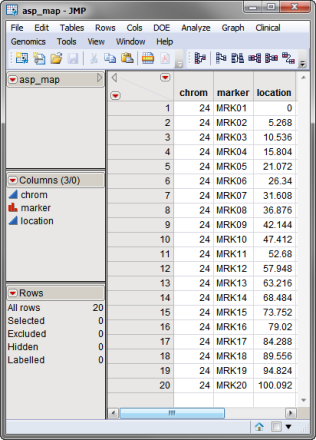
A second, optional, data set is the Annotation Data Set. This data set contains information such as gene identity or chromosomal location, for each of the markers. The annotation data set used in this example, the asp_map.sas7bdat data set, was computer generated and identifies each of the 20 markers in the IBD data set and their locations. This data set is illustrated below. This data set is a tall data set; each row corresponds to a different marker.
Both data sets are included in the Sample Data folder that comes with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the Affected Sib-Pair Tests output documentation for detailed descriptions and guides to interpreting your results.