Process Description
QTL IM, CIM and MIM Analysis
Interval Mapping (IM) is an extension of single marker analysis and it can do analysis on any genomic location flanked by two markers. IM uses complete marker linkage maps for genomic scanning (LR or LOD profile) for Quantitative Trait Loci (QTLs).
Composite Interval Mapping (CIM) is an extension of IM analysis that uses a combination of IM with multiple regression. CIM can improve the precision of QTL mapping in the situation of multiple QTLs in one chromosome.
The IM and CIM Analysis (Single QTL Model) process performs both types of mapping. Three SAS data sets are required: an Input Data Set of cross information, an Annotation Data Set that includes map information, and a genotype probability data set that includes probability information about the QTL genotypes.
What do I need?
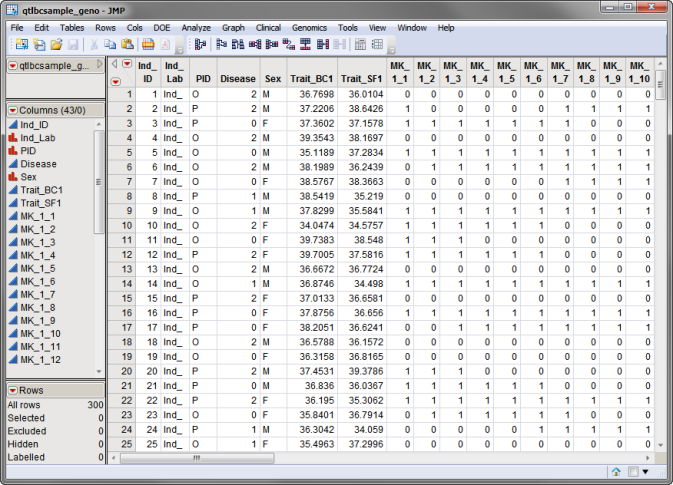
Three SAS data sets are required. The first, the input cross file, lists information about the different genetic crosses used in the study. The qtlbcsample_geno.sas7bdat data set, used in the following example, is shown below. This is a wide data set with 300 individuals listed in rows and the status of individuals for two traits and 36 markers, spanning 3 chromosomes, listed in columns. Markers are formatted as numeric genotypes.
The second required data set is an Annotation Data Set that lists map information for each of the markers. The qtlbcsample_anno.sas7bdat annotation data set, used in the following example, is shown below. This data set contains 4 columns listing the name and position (in cM) of 36 markers present on three chromosomes.

The third required data set is a genotype probability data set that lists the probabilities of observing specific QTL genotypes at each one cM interval along the chromosome(s) represented by the markers in the input data set. The qtlbcsample_geno_gp.sas7bdat genotype probability data set, shown below, lists probabilities for QTLs for the 300 individuals listed in the input data set (rows 3 through 302). Row 1 specifies the relevant chromosome, row 2 specifies the position (cM), and row 3 indicates markers (1 indicates marker and 0 inter-marker position).

The qtlsfsample_geno.sas7bdat, qtlsfsample_anno.sas7bdat, and qtlbcsample_geno_gp.sas7bdat data sets are located in the Sample Data\QtlMapping directory included with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the QTL IM, CIM and MIM Analysis output documentation for detailed descriptions and guides to interpreting your results.