Process Description
ArrayTrack Input Engine
The ArrayTrack Input Engine imports and combines your provided files from NCTR ArrayTrack into a SAS data set.
What do I need?
Before you can successfully import the raw data into SAS data sets that can be used for analysis in JMP Genomics, you must locate and gather several sources of information:
| • | An Experimental Design File (EDF) that indexes the individual .txt files for the experiment. The EDF is typically a text file or Excel spread sheet and must be created before the data can be imported. The AT_exp2.txt file shown below is one example of an EDF. |
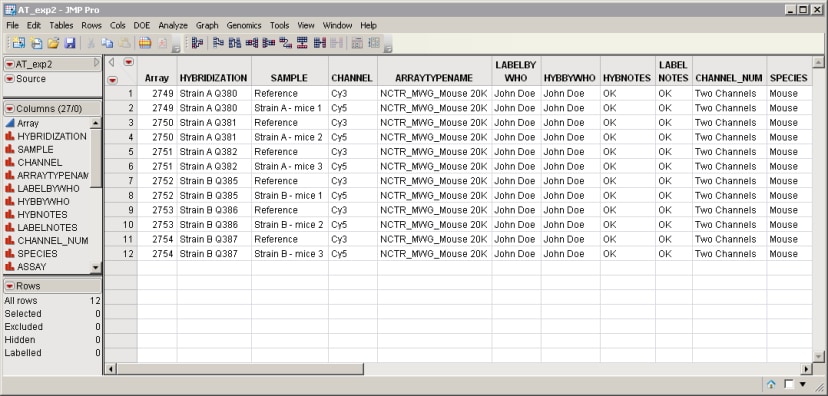
| • | The .txt files containing the raw data, which must be located and copied to a single folder. Each .txt file corresponds to an individual microarray, and contains the hybridization intensities and annotation information for that array. One such file is the AT_data2.txt file shown below. |

Both the AT_exp2.txt EDF and the AT_data2.txt file are located in the Sample Data\Microarray\ArrayTrack directory included with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output data sets generated by this process are listed in a Results window. Refer to the ArrayTrack Input Engine output documentation for detailed descriptions.