Process Description
Nanostring Input Engine
The Nanostring Input Engine imports a set of nanostring files and combines them into a single SAS data set.
What do I need?
Before you can successfully import the raw data into SAS data sets that can be used for analysis in JMP Genomics, you must locate and gather several sources of information:
| • | An Experimental Design File (EDF) that indexes the individual nanostring output Reporter Code Count (.RCC)files for the experiment. The EDF is typically a text file, Excel spread sheet, or SAS data set and must be created before the data can be imported. The edf.sas7bdat file shown below is one example of an EDF. |
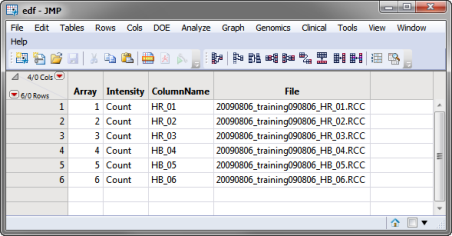
| • | All of the .RCC files containing the raw data must be located and copied to a single folder. RCC files are comma-separated text (.csv) files that contain the counts for each gene in a sample. The data for each sample hybridization are contained in a separate RCC file. One such file is the 20090806_training090806_HB_04.RCC file shown below: |
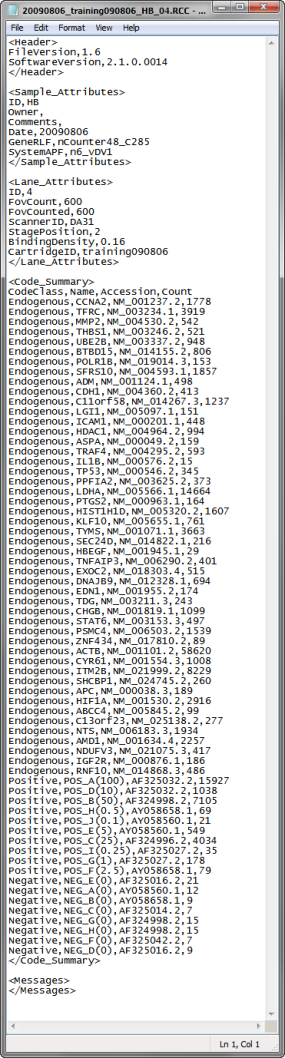
The file contains 4 comma separated variables: CodeClass, gene Name, gene accession number, and raw Count data.
Note: Because there might be slight differences in hybridization or purification, binding efficiency and other experimental variables should be normalized before comparing data between hybridizations.
The edf.sas7bdat experimental design file and the raw .RCC files are located in the Sample Data\Nanostring directory included with JMP Genomics.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output data sets generated by this process are listed in a Results window. Refer to the Nanostring Input Engine output documentation for detailed descriptions.