Process Description
2D Detrend
Spectral data often contain an unwanted baseline trend that varies from spectrum to spectrum. Removing such trends is recommended to ensure comparability of the spectra.
The Spectral 2D Detrend process removes a baseline trend from each member of a group of two-dimensional spectra in order to make them statistically comparable.
What do I need?
One data set is required to run the Spectral 2D Detrend process. This data set must be in tall format, with one variable (column) for each spectrum and another variable for the value of the m/z or time-of-flight index. The name of the input SAS data set must be lowercase.
The wright_tall_2k_10k.sas7bdat data set, shown below, lists 165 individual spectra. The mz column lists the m/z values for each of the points in the spectra.
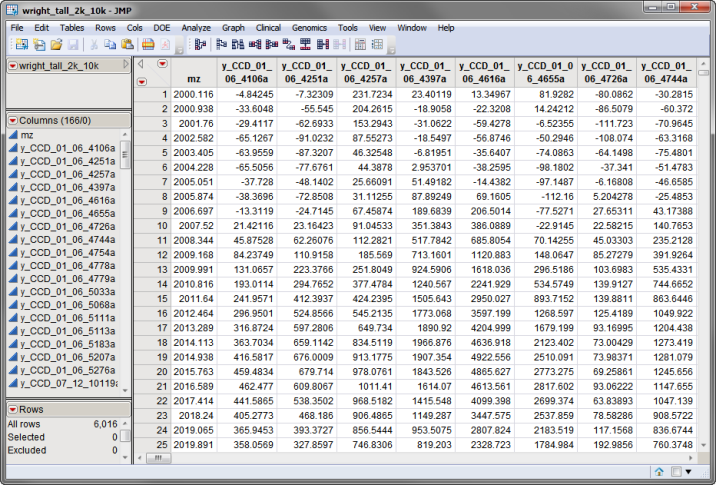
The wright_tall_2k_10k.sas7bdat data set is located in the Sample Data\Proteomics directory.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
Refer to the 2D Detrend output documentation for detailed descriptions of the output and guides to interpreting your results..