Process Description
Stack
SAS data sets are typically formatted in one of two ways: wide or tall. In wide data sets, samples are listed in columns, and molecular entities, such as genes or proteins, are listed in rows. Tall data sets are the transpose of wide data sets, with molecular entities listed in columns and samples listed in rows. A tall data set has one or more common feature identifiers (ID column(s)) across columns of interested measurements. Typically, the number of rows is greater than the number of columns in this format of data sets. Analyses of tall data sets typically also require an additional Experimental Design Data Set (EDDS). Different procedures in JMP Genomics have different requirements for data set format. Please refer to Data Sets Used in JMP Genomics Processes for a more detailed description of these data set formats.
Many SAS procedures ( PROCs), however, require that input data sets be formatted in a stacked format. A stacked data set has all of the observations of interest stacked into a single variable, or column. Rows are organized into groups of similar observations in which each row in the group differs in only one experimental parameter. Each group differs from other groups in additional parameters. A stacked data set typically has many, many more rows than columns.
The Stack process combines and restructures a tall formatted SAS data set with an accompanying EDDS into a stacked format. This process enables you to quickly get your data into a stacked format for directly running (or testing) SAS processes.
What do I need?
Two data sets are required for this process:
| • | An Input SAS Data Set (see Input Data Set) in tall format. |
| • | An Experimental Design Data Set. This data set tells how the experiment was performed, providing information about the columns of the primary experimental data. See Experimental Design Data Set (EDDS) for more information. |
The affylatin_norm.sas7bdat data set (found in the \LifeSciences\Sample Data\Microarray\Affymetrix Latin Square directory included with JMP Genomics and JMP Clinical, and described in Affymetrix Latin Square Data) serves as an example Input SAS Data Set.
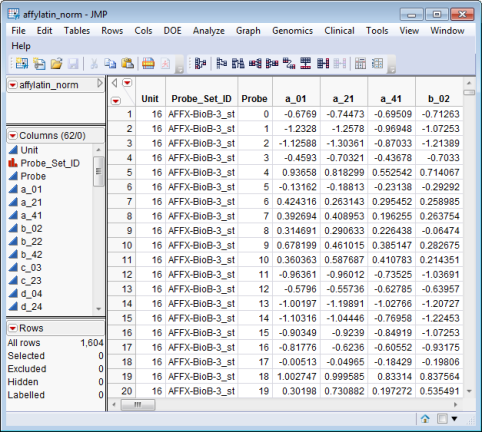
The affylatin_exp.sas7bdat data set (found in the \LifeSciences\Sample Data\Microarray\Affymetrix Latin Square directory included with JMP Genomics and JMP Clinical, and described in Affymetrix Latin Square Data) serves as an example Experimental Design SAS Data Set.
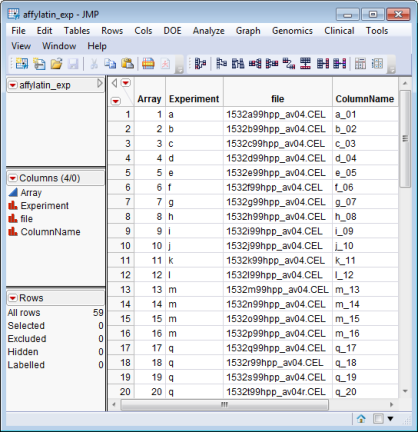
The file and ColumnName columns are to be excluded from the Output Stack Data Set.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output of the Data Stack process includes a Results window, which lists the name of and path to the Output Stack Data Set. To best visualize this data set, think of the Experimental Design SAS Data Set as a scale diagram used to guide the unpacking of the “boxy” 1604x59-data cell structure (the intensity data in the Input SAS Data Set) into a 94636x1 tower.
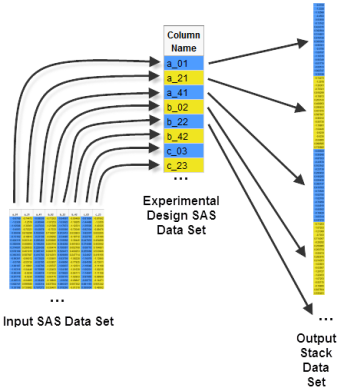
The affylatin_norm_stk.sas7bdat Output Stack Data Set is shown below.

Notice the immense increase in row count, and decrease in column count.
This data set is ready for further analysis.
See also
| • | Unstack |