Running this process for the
GeneticMarkerExample
sample setting generates one output data set accessed from a
Results
window shown below. Refer to the
Recode Missing Genotypes
process description for more information about this process.
The
Results
window contains the following panes:
|
•
|
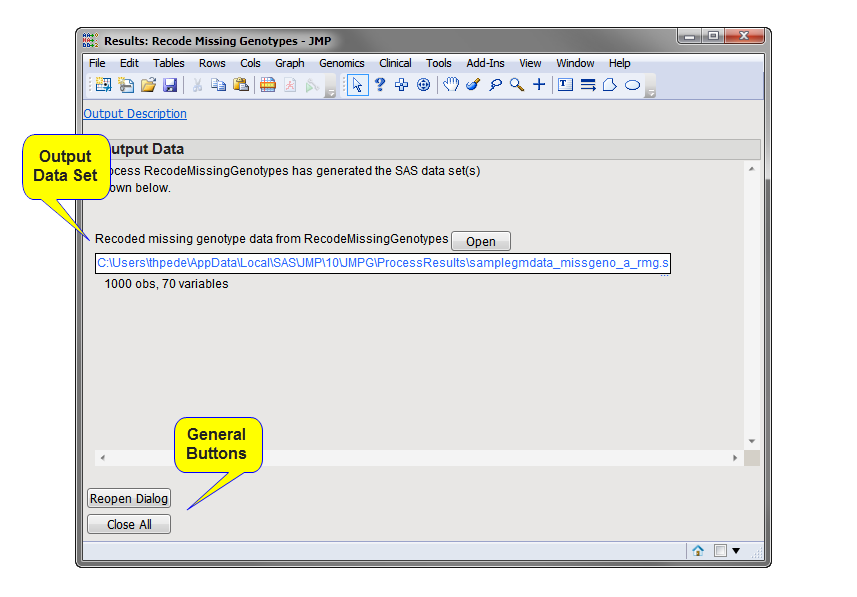
Output Genotype Data Set:
The
output data set containing the
genotype
data from the input data set recoded as specified in the process
dialog
. In this example, the missing character genotypes, represented in the input data set by
asterisks
(
*
), were replaced with the
blanks
(
) required by JMP Genomics genetic processes for missing character genotypes. Compare the genotypes at markers
g1
and
g2
for the first 10 individuals (shown below).
|
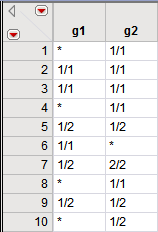
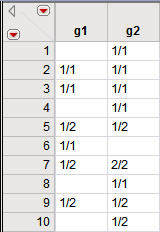
The panel on the left, which comes from the input data set, shows the genotypes for the first ten individuals at markers
g1
and
g2
. Two
alleles
delineated by a “/” are present at these loci, with allele "1" being the most common. The panel on the right, which comes from the output data set, shows the same data. Missing genotypes are indicated by blank spaces.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
|
•
|
Click
to reopen the completed process dialog used to generate this output.
|
|
•
|
Click
to close all graphics windows and underlying data sets associated with the output.
|