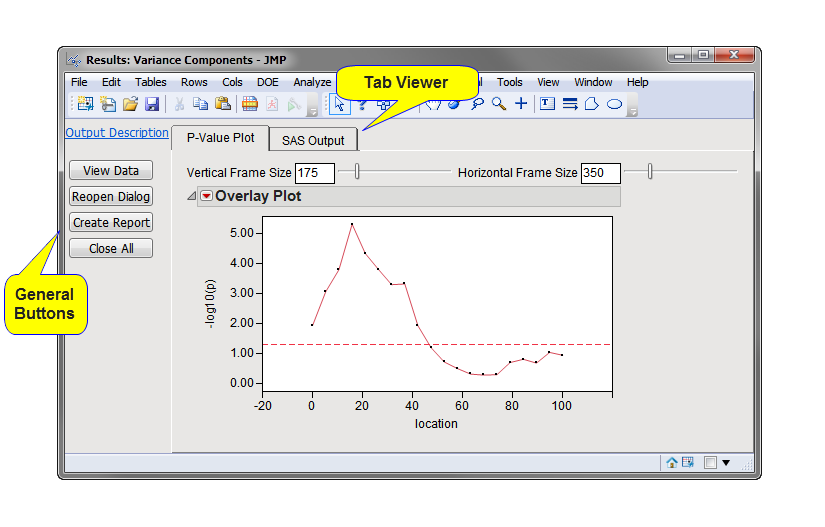
Running this process using the
Merlin_asp
sample setting generates the tabbed
Results
window shown below. Refer to the
Variance Components
process description for more information. Output from the process is organized into tabs. Each tab contains one or more plots, data panels, data filters, and so on. that facilitate your analysis.
This pane provides you with a space to view individual tabs within the
Results
window. Use the tabs to access and view the output plots and associated data sets.
|
•
|
Summary Chart
: When there are multiple annotation groups (
chromosomes
or genes, for example), this tab displays the number of significant markers in each annotation group for each test.
|
|
•
|
Manhattan Plot
: When there are multiple annotation groups (chromosomes or genes, for example), this tab displays a scatter plot of the
p-values
across all annotation groups.
|
|
•
|
Annotation Group Results
: When there are multiple annotation groups (chromosomes or genes, for example), a separate
Results
tab with an
overlay plot
of
p
-value by chromosome location is created for each annotation group. In this example, there are no annotation groups specified.
|
|
•
|
All P-Value Plots
: When there are multiple annotation groups (chromosomes or genes, for example), the
All P-Value Plots
tab shows all the
p
-value plots from the
Annotation Group Results
tabs in a single display.
|
Note
: When an annotation group
variable
is not specified, as in this example, or there is only one annotation group, the tab is named
P-Value Plot
and contains an
overlay plot
of
p
-value by chromosome location for all markers.
|
•
|
SAS Output
: This tab displays the SAS output from running PROC MIXED to fit the
variance
components model, including one with the null model including
covariates
only then a run of PROC MIXED for each marker for the alternative model.
|
|
•
|
GenBank Nucleotide
: Select points or rows and click
to opens a browser window directed to a GenBank Nucleotide search for the selected molecular entities.
|
|
•
|
UniGene Database
: Select points or rows and click
to access information from the Unigene database for selected genes or markers
|
|
•
|
AceView Database
: Select points or rows and click
to access information from the ACEView database for selected genes or markers.
|
|
•
|
|
•
|
Click
to reveal the underlying data table associated with the current tab.
|
|
•
|
Click
to reopen the completed process dialog used to generate this output.
|
|
•
|
Click
to generate a
pdf
- or
rtf
-formatted report containing the plots and charts of selected tabs.
|
|
•
|
Click
to close all graphics windows and underlying data sets associated with the output.
|