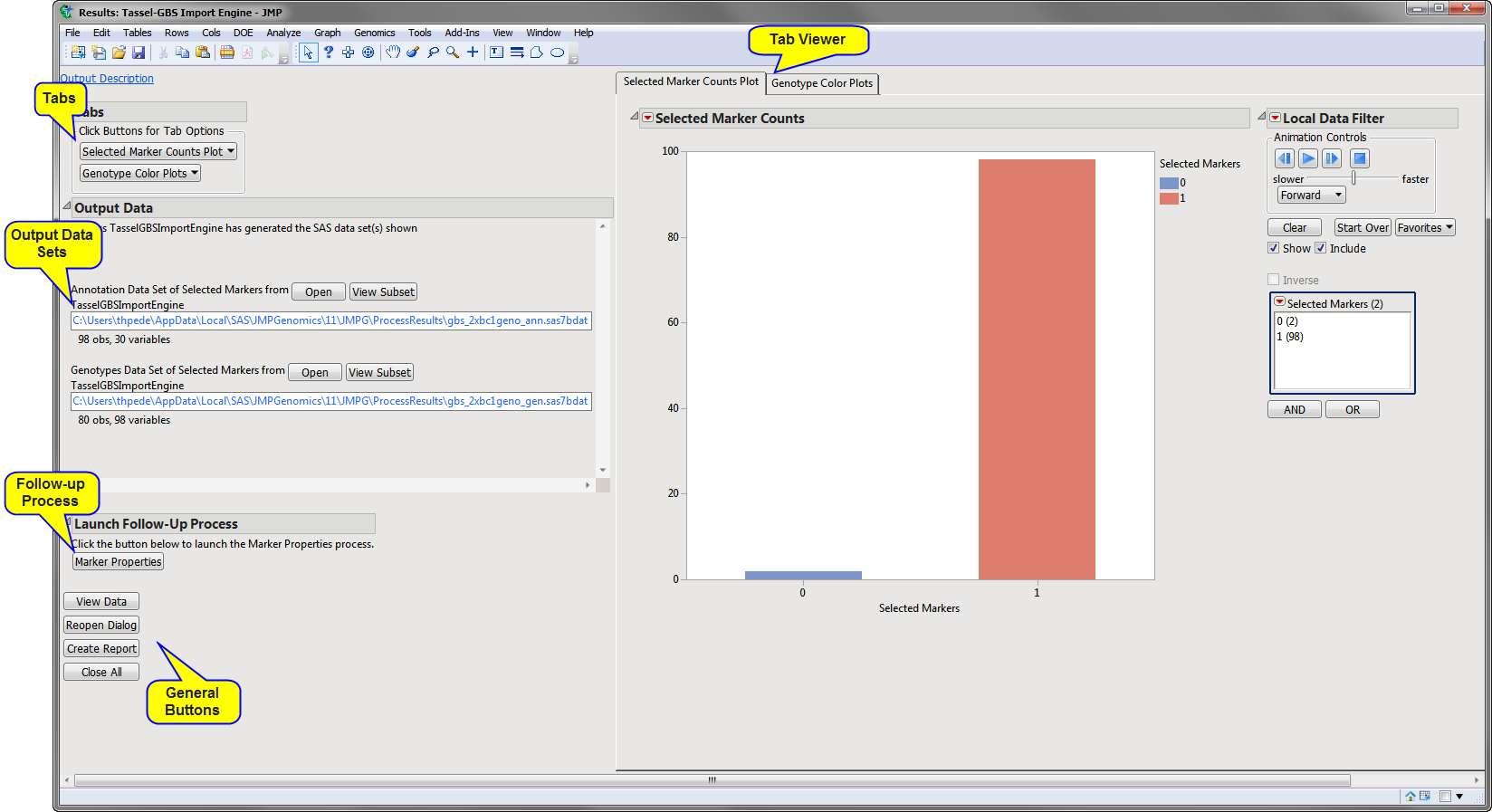
Running this process for the
gbs_2xbc1geno
sample setting generates the
Results
window shown below. Refer to the
Tassel-GBS Import Engine
process description for more information.
The
Results
window contains the following panes:
This pane provides you with a space to view individual tabs within the
Results
window. Use the tabs to access and view the output plots and associated data sets.
|
•
|
Selected Marker Counts Plot
: Shows a plot of selected marker counts.
Note
: This tab is generated only when
Outcross
in
NOT
selected as the
Cross Type
.
|
|
•
|
Genotype Color Plots
: This tab displays a
cell plot
for each individual (in the rows) colored by the
genotypes
of markers (in the columns). These plots are useful to detect potential genotyping errors in the data and genotype messiness.
Note
: This tab only is displayed when the
Display marker genotype cell color plots
check box option is checked.
|
|
•
|
Marker Type Counts Plot
: This tab that shows a plot of selected markers counts for each cross type from a bi-parental cross of outbred parents.
Note
: This tab is generated only when
Outcross
is selected as the
Cross Type
.
|
|
•
|
P-Value Plots of Test I
: This tab shows the raw
p-values
of a
chi-square
test that is conditional on the genotypes of parents.
Note
: This tab is generated only when
Outcross
is selected as the
Cross Type
.
|
|
•
|
P-Value Plots of Test II
: This tab shows the raw
p
-values of a
chi
-square test that is
NOT
conditional on the genotypes of parents.
Note
: This tab is generated only when
Outcross
is selected as the
Cross Type
.
|
|
•
|
Genotypes Data Set of Selected Markers
: This data set contains the
Marker ID Variable
,
Parent 1 ID Variable
,
Parent 2 ID Variable
,
Individual Variables
, and
Variables to Keep in Output Data Set
that were specified on the
General
tab. Columns of relevant summary statistics as well as coded genotypes for each selected marker are also included.
Variables
that have
_testI
or
_testII
in their names come from the
Chi
-square Test I or
Chi
-square Test II, respectively.
Note
: The
Chi
-square Test I is conditional on the genotypes of parents, the
Chi
-square Test II is not.
|
|
•
|
Annotation Data Set of Selected Markers
: This data set contains relevant columns of summary statistics for the subset of markers selected from the analyses, plus all columns from the
Annotation SAS Data Set
(provided one has been specified. Variables that have
_testI
or
_testII
in their names come from the
Chi
-square Test I or
Chi
-square Test II, respectively. The
Chi
-square Test I is conditional on the genotypes of parents, the Chi-square Test II is not.
|
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
|
•
|
Marker Properties
: Click
to launch the
Marker Properties
process with the Genotypes and
annotation data sets
specified as input.
|
|
•
|
Click
to reveal the underlying data table associated with the current tab.
|
|
•
|
Click
to reopen the completed process dialog used to generate this output.
|
|
•
|
Click
to generate a
pdf
- or
rtf
-formatted report containing the plots and charts of selected tabs.
|
|
•
|
Click
to close all graphics windows and underlying data sets associated with the output.
|