|
|||||
|
|||||
|
|||||
|
|||||
Note
: For
METHOD=MSPL
,
METHOD=MMPL
,
METHOD=LAPLACE
, and
METHOD=QUAD
, the
IC=Q
and
IC=PQ
options produce the same results.
|
|||||
Note
: For
METHOD=MSPL
,
METHOD=MMPL
,
METHOD=LAPLACE
, and
METHOD=QUAD
, the
IC=Q
and
IC=PQ
options produce the same results.
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
|||||
|
Note
: In generalized linear models with normally distributed data, you can use the PROFILE option to request profiling of the residual variance.
|
|||||
|
|
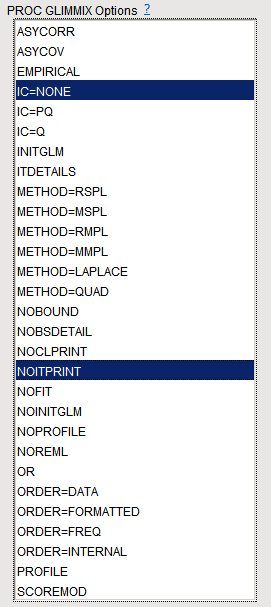
To specify more than one option, hold down
as you left-click on the desired options.
|
Refer to the
SAS PROC GLIMMIX documentation
for more information and references.