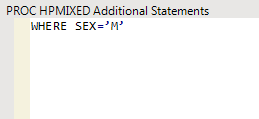|
•
|
PARMS for specifying initial values of the
covariance
parameters or performing a grid search of over several values of these parameters.
|
Refer to the
SAS PROC HPMIXED documentation
for appropriate syntax.
|
•
|
Option
is the PROC HPMIXED option, and
|
|
•
|
a
space
is used to delimit individual conditions.
|
|
Most
models
fit with the GLIMMIX procedure typically have one or more nonlinear parameters. Estimation requires nonlinear optimization methods. You can control the optimization through options using the
NLOPTIONS
statement.
See the
NLOPTIONS Statement documentation
of more information
|
|
|
The PARMS statement specifies initial values for the covariance or scale parameters, or it requests a grid search over several values of these parameters in generalized linear
mixed model
.
See the
PARMS Statement documentation
for more information.
|
By default,
CLASS
,
MODEL
,
RANDOM
, and
OUTPUT
statements are automatically generated to fit the Genomic BLUP model, so do
not
specify any of these statements. Also, do
not
specify a
BY
statement, since the SAS code is currently not set up to handle
BY groups
.
|
|
Type specific PROC HPMIXED options in the
PROC HPMIXED Additional Statements
field.
|
For example, to restrict the analysis to males only, type
WHERE SEX=’M’
in the text field, as shown below:
Refer to the
SAS PROC HPMIXED documentation
for more information.