The
Batch Normalization
process normalizes data by establishing a batch profile based on averaging across within-batch-level control arrays and then using this profile to correct values across all arrays.
K-Means clustering
is applied for grouping batch profiles into clusters. The batch
normalization
is performed by correcting the within-cluster
mean
profile for each cluster. Standardizing for batch is an option before clustering batch profiles.
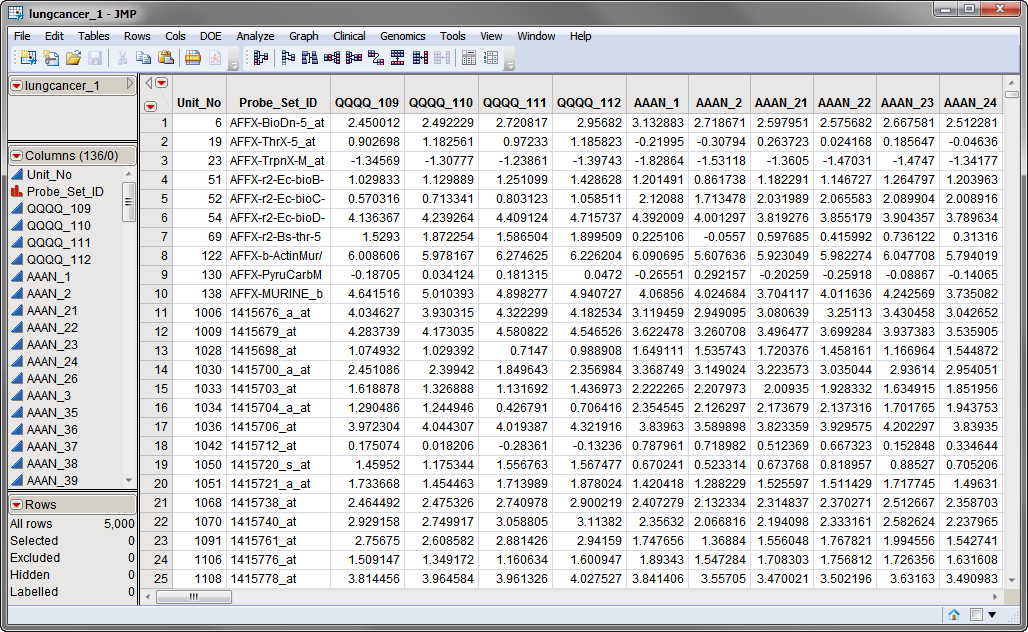
The first data set, the
Input Data Set
, contains all of the numeric data to be normalized.
The
lungcancer_1.sas7bdat
data set, shown below, represents data collected in multiple independent studies over the course of several years. A total of 134 arrays representing animals treated or not treated (Control) with various dosages of different potentially anti-cancer agents and assessed for gene
expression
using 5000 different
probesets
. Data was collected from four different studies.
Note that this is a
tall
data set; each
probe
corresponds to one row whereas each column corresponds to a separate experimental condition.
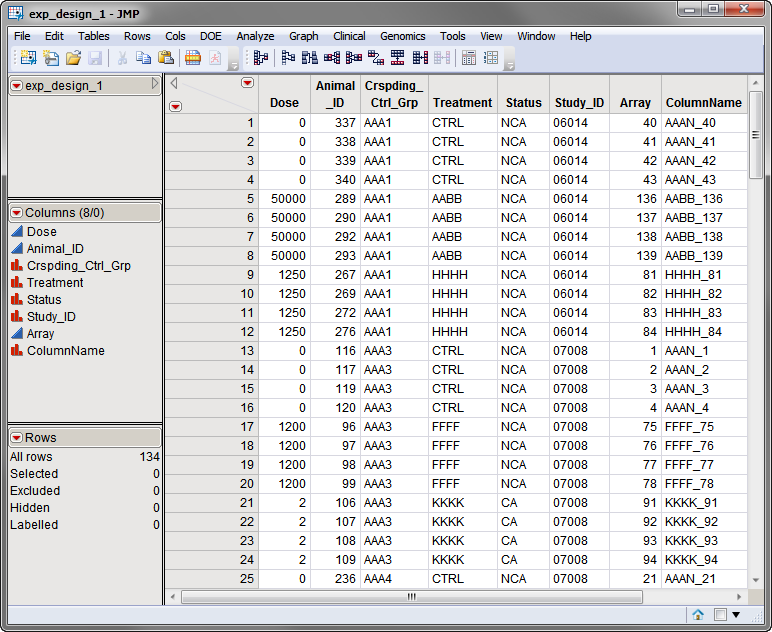
The second data set is the
Experimental Design Data Set (EDDS)
. The
exp_design_1.sas7bdat
EDDS is shown below. This required data set tells how the experiment was performed, providing information about the columns in the input data set. Note that one column in the EDDS must be named
ColumnName
and the values contained in this column must exactly match the column names in the input data set.
The
lungcancer_1.sas7bdat
data set and
exp_design_1.sas7bdat
experimental design data set are included in the
Sample Data\Affymetrix Lung Cancer
folder.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
The output generated by this process is summarized in a Tabbed report. Refer to the
Batch Normalization
output documentation for detailed descriptions and guides to interpreting your results.