Genotypic information for sets of
alleles
or markers is frequently listed using combinations of letters and numbers. Alternatively, data sets can use a two column
allelic
format in which each of the two alleles that make up a
genotype
is listed in a separate column. Such formats are not amenable to many forms of statistical analysis, which typically can use only single-column, numerical data. Before any analyses can be performed. Therefore, data sets containing genetic marker data in a non-numerical and/or allelic format must be recoded. Conversely, genotypes might have been originally coded numerically as the values 0, 1, and 2 or the genotype might be coded as a single letter -- "A" for genotype A/A, "B" for genotype B/B, and "H" for the
heterozygous
genotype A/B. Since the processes for analyzing genotype data require a representation that discerns the two alleles that the genotype comprises, converting the single character or numeric genotypes to this recognized form is necessary.
When numeric recoding of genotypes is performed,
Recode Genotypes
tabulates the
SNP
genotypes for all of the individuals in a data set, ranking the prevalence of each allele. Depending on the recoding desired, the process assigns numerical identities to the genotypes based on the prevalence of the alleles, whether they are
homozygous
or heterozygous, and their dominant/recessive relationship. Conversely,
Recode Genotypes
can also convert single character numeric (0, 1, and 2) or non-numeric (A, B, and H) back into a genotypic string (A/A, B/B, and A/B).
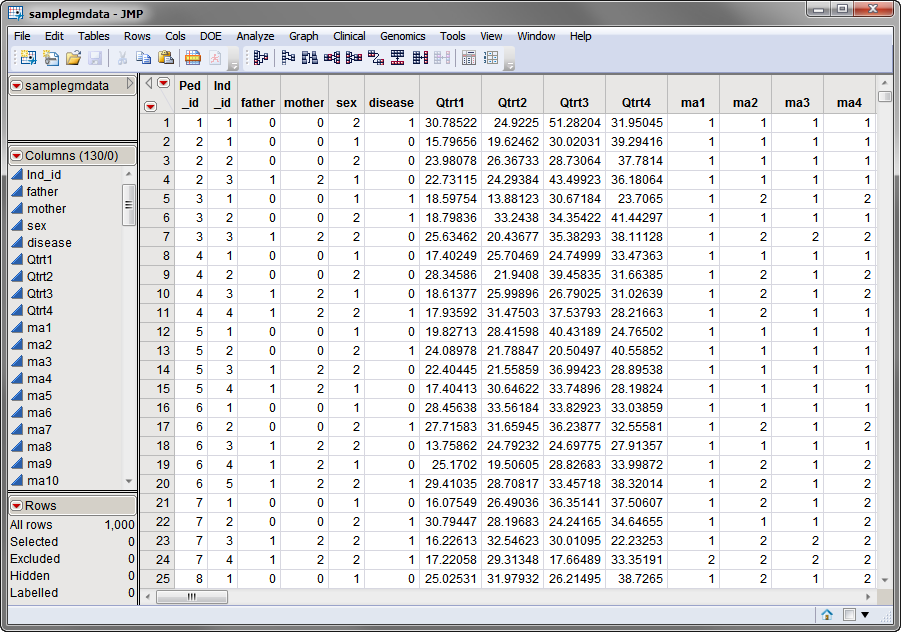
Two data sets are needed for this process. The first, the
Input Data Set
, contains all of the marker data. The sample data set used in the following example, the
samplegmdata
data set, was computer generated as described in
Sample Genetic Marker Data
. It consists of 1000 rows of individuals with 130 columns corresponding to data on these individuals. Marker data is presented in the two-column format. This data set is partially shown below. Note that this is a wide data set; markers are listed in columns, whereas individuals are listed in rows.
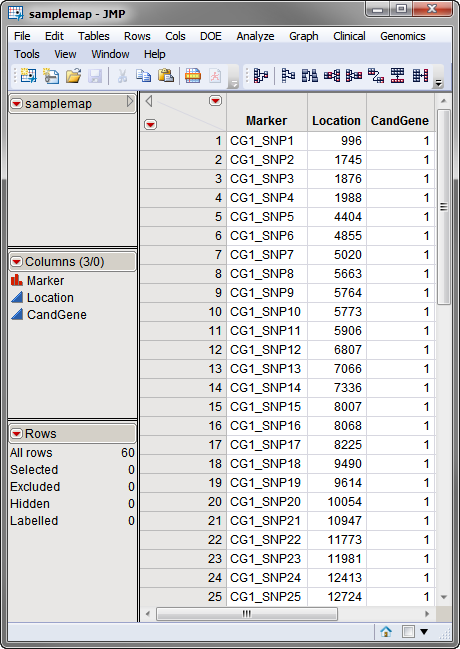
The second data set is the
Annotation Data Set
. This data set contains information, such as gene identity or chromosomal location, for each of the markers. The
annotation data set
used in this example, the
samplemap
data set, was computer generated and identifies markers, location and gene identities. A portion of this data set is illustrated below. This data set is a tall data set; each row corresponds to a different marker.
Note
: The top-to-bottom order of the rows in the annotation data set matches the left-to-right order of the columns in the input data set. This correspondence is required for this process.
Both data sets are included in the
Sample Data
folder.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
Output from this process is accessed from a
Results
window. Refer to the
Recode Genotypes
output documentation for detailed descriptions and guides to interpreting your results.