The
SAM Input Engine
imports a set of Sequence Alignment Map (
.sam
) files and combines them into three SAS data sets containing
chromosome
, location, and sequence identity of each sample with a reference sequence.
Note
: This process is considered
experimental
.
|
•
|
An
Experimental Design File (EDF)
that indexes the individual raw data files for the experiment. The EDF is typically a text file or Excel spread sheet and must be created before the data can be imported.
|
Important
: The EDF used for importing SAM data must contain a
variable
called
SampleName
. The values listed in this column identify each sample in the experiment. Providing this name allows the SAM data for each sample, which are typically spread across multiple
.sam
files, to be merged into one SAS data set.
|
•
|
All of the
.sam
files containing the raw data, which must be located and copied to a single folder. Each
.sam
file corresponds to an individual chromosome and contains the hybridization intensities for that array.
|
The following example uses
.sam
files for the Y chromosome of two different mice (from the list shown below) that were downloaded from
The 1000 Genomics Project
and saved in the
Next-Gen\SAM\GSE18905
folder created in the JMP Genomics
Sample Data
folder.
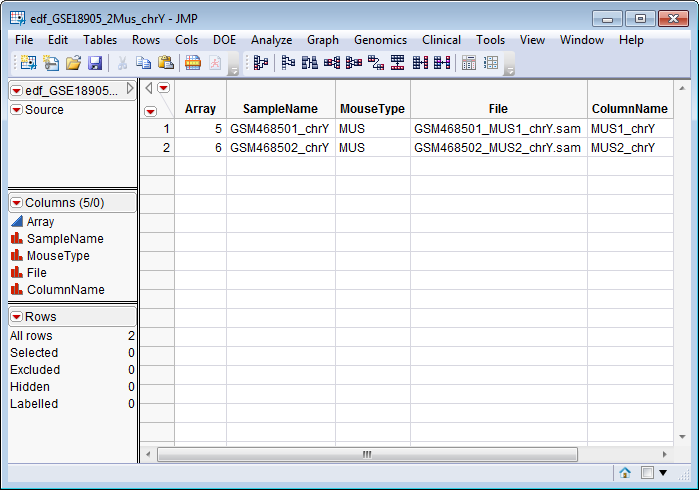
The EDF for this example (shown below) specifies the import of the
GSM468501_MUS1_chrY.sam
and
GSM468501_MUS2_chrY.sam
files.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
The output data sets generated by this process are listed in a
Results
window. Refer to the
SAM Input Engine
output documentation for detailed descriptions.