The
Subset
process subsets an input data set based on the criteria specified by up to four different filters.
For example, consider the
Drosophila
Aging experiment described in
Drosophila Aging Experimental Data
. The
drosophilaaging_exp.sas7bdat
data set (found in the \
LifeSciences\Sample Data\Microarray\Scanalyze Drosophila
directory included with JMP Genomics), shown
below
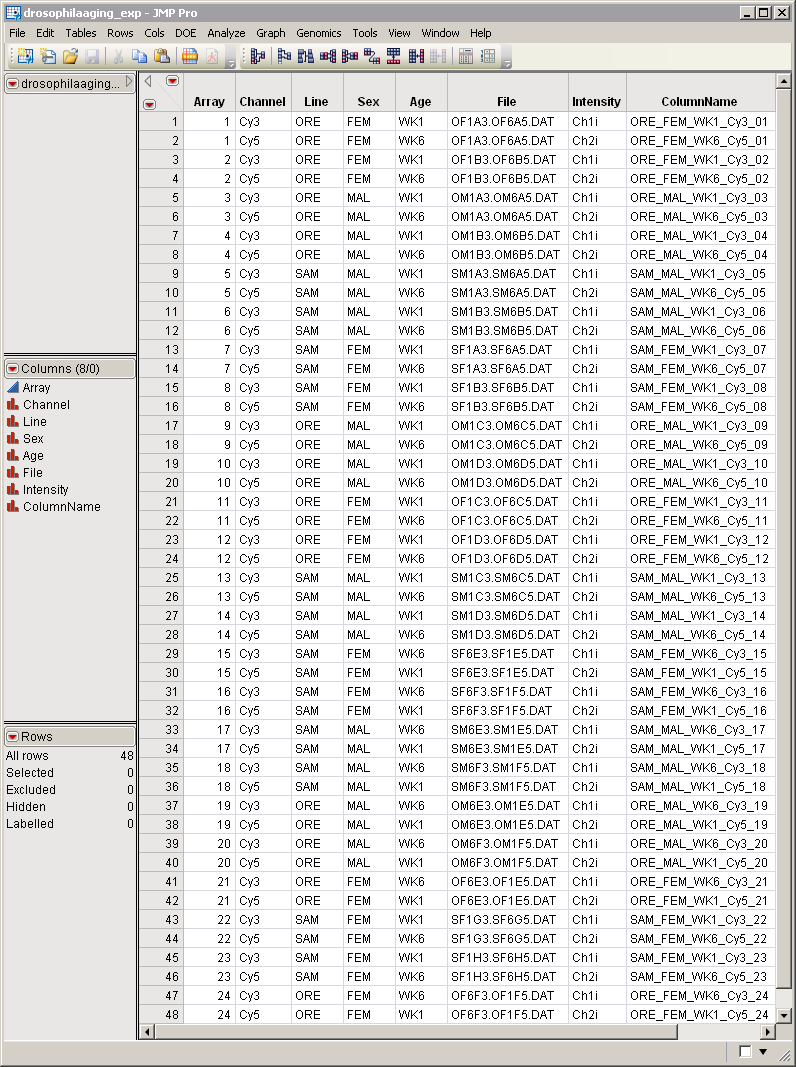
, serves as the input data set. This experimental design data set contains information for the
Drosophila
Aging experiment and is used to identify and describe files containing raw data. The data set contains eight columns and 48 rows.
In this example, we generate a subset of the
drosophilaaging_exp.sas7bdat
data set containing only those rows specifying data from female flies from specific arrays (all except array “1”). Since all data are from female flies, we also drop the
variable
specifying the sex of the flies.
Make the following choices in the
dialog
to follow this example:
|
|
|
|
|
|
|
|
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
|
|
Click
to view the
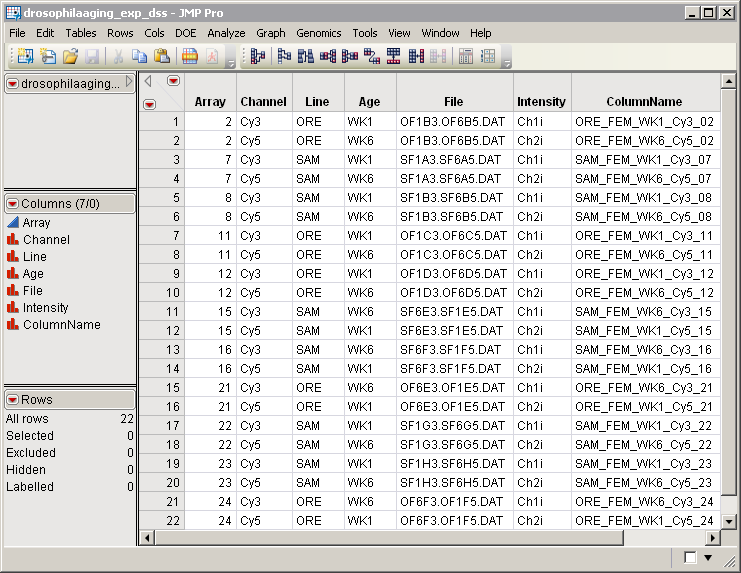
drosophilaaging_exp_dss.sas7bdat
output data set.
|
The output data set contains 7 columns and the 22 rows. Only the rows specifying data for female flies have been included. The
Sex
column has been excluded. All rows for array
1
have been excluded.