The
Workflow Builder
takes advantage of JMP Genomics' ability to save settings (see
Saving and Loading Settings
) to enable you to string together groups of processes into a comprehensive analysis series. A typical, well-designed
workflow
can take you all the way from the initial data import, through preliminary analysis, quality control, statistical analysis, modeling and annotation of results. The workflow series can be saved for use again and again for different data sets, so long as the basic experimental design remains constant. Existing workflows can also be edited as desired to accommodate changes in experimental design.
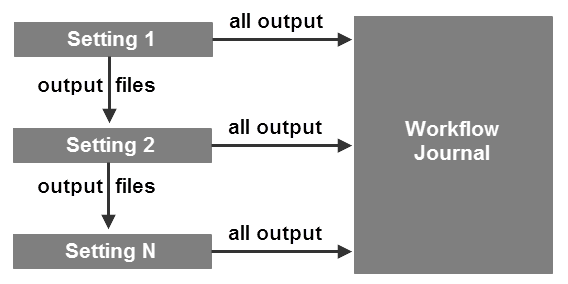
Each setting consists of a process, along with its associated parameters (input filenames, specific options, and output filenames). The output files from each setting are used as input for each subsequent setting. The complete output (graphical, tabular, and file) from each setting is written to the workflow
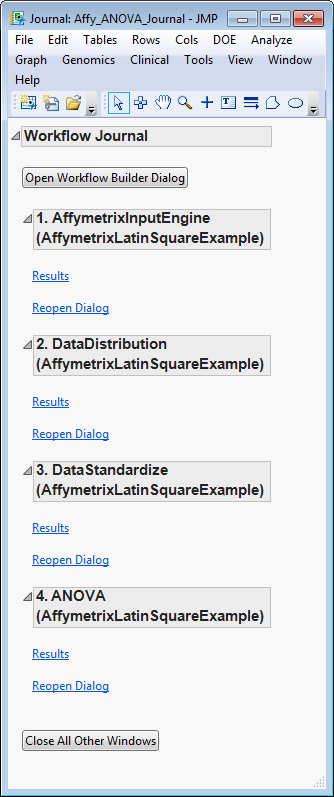
journal
. Thus, the workflow journal contains output from the entire workflow.
|
•
|
which
specific processes you want to run as a group, and
|
|
•
|
Each process needs its own
Input Data Set
, which might or might not have been created by a previous process in the workflow. If the workflow requires an import of raw data, the appropriate data import engine as well as each of the processes to be used in the workflow must be configured, and the settings saved in
one
settings folder. Finally, an
output folder
must be created, into which all of the resulting data sets, analyses, graphics, and other output are placed.
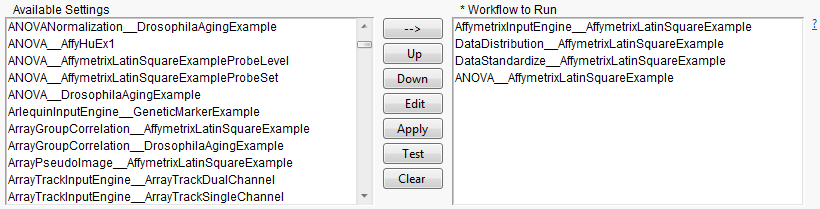
A sample workflow, in the process of construction, is shown below. Using the selection and ordering buttons, the workflow currently starts with
Affymetrix Input Engine
, which is followed by
Data Distribution
, which is then followed by
Data Standardize
, and ends with
ANOVA
. The output from each process becomes input for each subsequent process.
The
Workflow Builder
is not intended for first time users. Instead, it was designed for power users who have already explored the many options offered through JMP Genomics. It is assumed that you are familiar with the processes, and have settled upon one or more standard protocols for your analyses. You also typically have specific saved settings for each of the processes.
Tip
: When saving settings for individual processes, you can now opt to save those settings to a newly saved workflow by checking the
Save to Workflow
box. If the
Workflow Builder
is open, the new setting is added to the current workflow. If the
Workflow Builder
is
not
open, saving a new setting with this box checked launches the
Workflow Builder
with the newly saved setting in place.
The set of saved settings that comprise a workflow can be edited either in the
dialog
for that process or in the
Workflow Builder
itself. If you are not familiar with the individual processes that you want to use, consult the specific documentation for those
Processes
for more information.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see
Files and Data Sets
.
Tip
: As you become more experienced with workflows, you can apply them directly to new experiments. In this case, it is suggested that you use generic names for all input and output data sets, and use folder names to identify the experiments. That way, the results will not contain data set names from previous experiments.