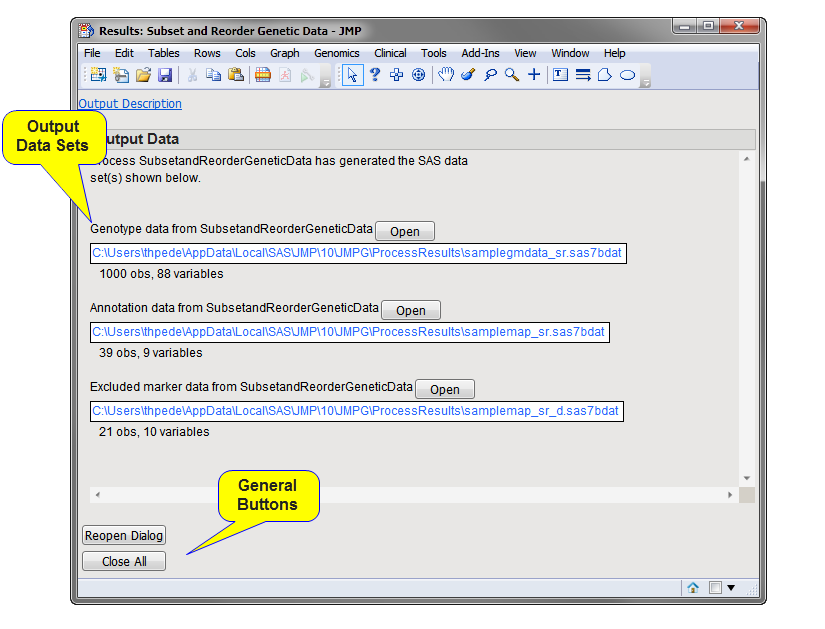
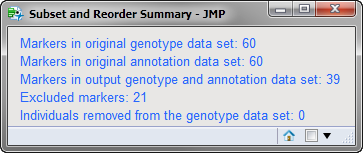
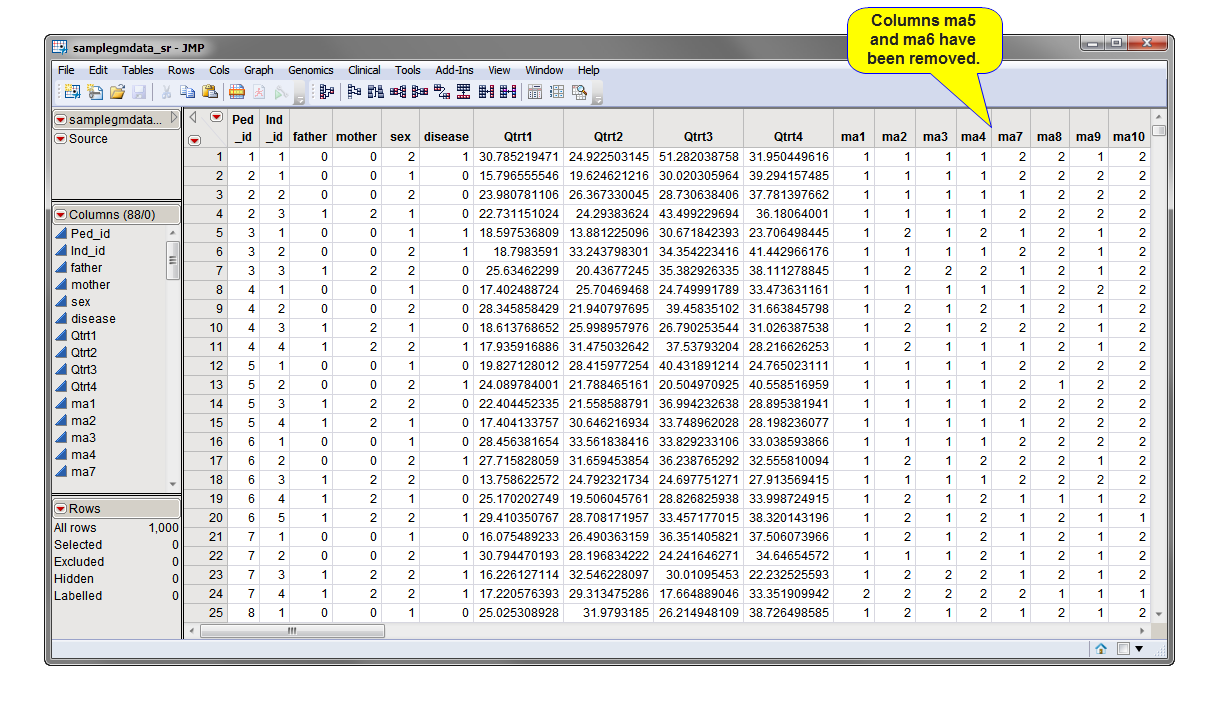
Running this process for the GeneticMarkerExample sample setting generates one output data set accessed from the Results window shown below. Data in the MAF (minor allele frequency) column, contained in the annotation data set, was used to subset the markers so that only those markers with a minor allele frequency greater than 0.1 were included in the output data. Refer to the Subset and Reorder Genetic Data process description for more information about this process.
The Results window contains the following panes:
|
•
|
Genotype Data Set: The output data set consists of all of the subsetted and reordered data. Click to view the output genotype data set.
|
|
•
|
Annotation Data Set: This data set contains the annotation data for the subsetted markers in the output data set.
|
|
•
|
Excluded Marker Data Set: This data set lists summary statistics for the excluded markers.
|
The output data set and annotation data sets are both denoted by the appended _sr suffix. The excluded marker data set is denoted by the _sr_d suffix. For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
|
•
|
Click to reopen the completed process dialog used to generate this output.
|
|
•
|
Click to close all graphics windows and underlying data sets associated with the output.
|