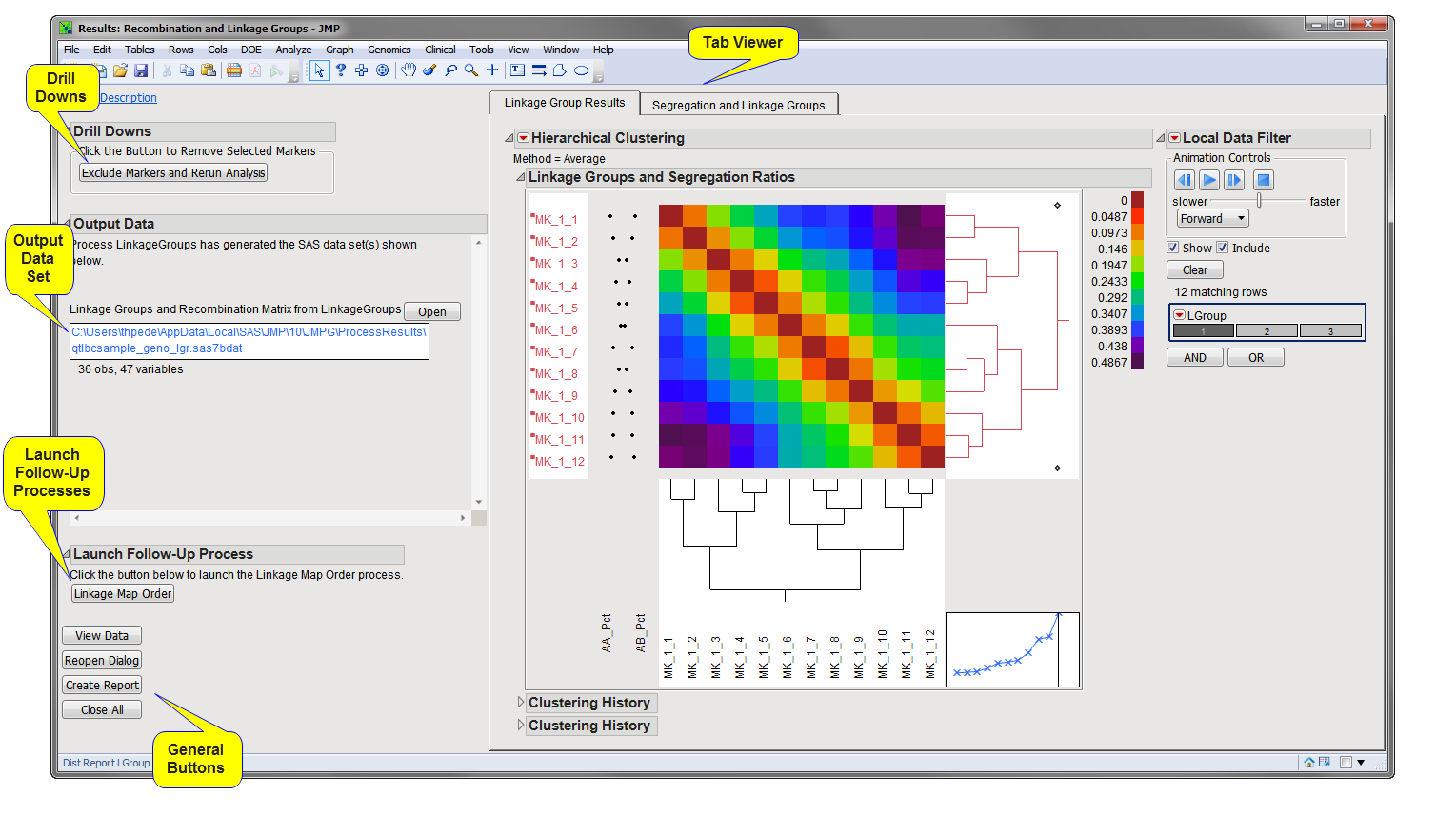
Running this process using the BC1_example sample setting generates the tabbed Results window shown below. Refer to the Recombination and Linkage Groups process description for more information. Output from the process is organized into tabs. Each tab contains one or more plots, data panels, data filters, and so on. that facilitate your analysis.
This pane provides you with a space to view individual tabs within the Results window. Use the tabs to access and view the output plots and associated data sets.
|
•
|
Linkage Group Results: This tab displays the recombination matrix and linkage group assignment for markers in your experimental population using the JMP Hierarchical Clustering Platform. This tabbed pane will be shown only if the Plot heatmaps for linkage groups option is checked.
|
|
•
|
Linkage Groups and Segregation: This tab displays a plot of p-values from testing segregation distortion each marker and distribution plot(s) of markers belonging to each linkage group as well as markers belonging to Framework Map groups (if a Framework Map was specified).
|
Note: This tab will not be shown if AIL or IRIL is selected as the Cross Type and no values have been entered for the Expected Segregation Ratios for AA AB BB parameter on the dialog.
|
•
|
Save Current Linkage Group Membership: Click and hold the diamond (
|
Note: This action button is available only when you choose the Interactive Hierarchical Clustering linkage group method.
|
•
|
Exclude Markers and Rerun Analysis: Click to rerun recombination and linkage grouping while excluding any selected markers.
|
|
•
|
Linkage Groups and Recombination Matrix from Linkage Groups: This is output data set contains the computed recombination matrix between all markers and the linkage group assignment. This data set is ready to be used for the Linkage Map Order process to continue with linkage map construction. This data set contains any interactive results, unless the option to Plot heatmaps for linkage groups check box is unchecked.
|
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
|
•
|
Linkage Map Order: Click to launch the Linkage Map Order process with the output linkage groups and recombination data set and the original input genotype data set preloaded as input.
|
|
•
|
Click to reveal the underlying data table associated with the current tab.
|
|
•
|
Click to reopen the completed process dialog used to generate this output.
|
|
•
|
|
•
|
Click to close all graphics windows and underlying data sets associated with the output.
|