An interesting part of any genomics analysis occurs after you have completed the statistical analysis and identified one or more significant genes. What functional roles do the genes play? How are they related to each other? In what biochemical pathways are they involved? JMP Genomics provides you with a set of bioinformatic tools that can help address these questions and incorporate biological meaning into your statistical results.
The Ingenuity Pathways Analysis Upload process creates an .html-formatted export file that can be uploaded to the Ingenuity Pathways Analysis (IPA) system for contextual analysis of expression and functional data for a suite of genes under specific experimental conditions.
One data set is required to run this process. This data set should include the results of a prior JMP Genomics analysis and must contain IPA-supported gene or protein identifiers and variables.
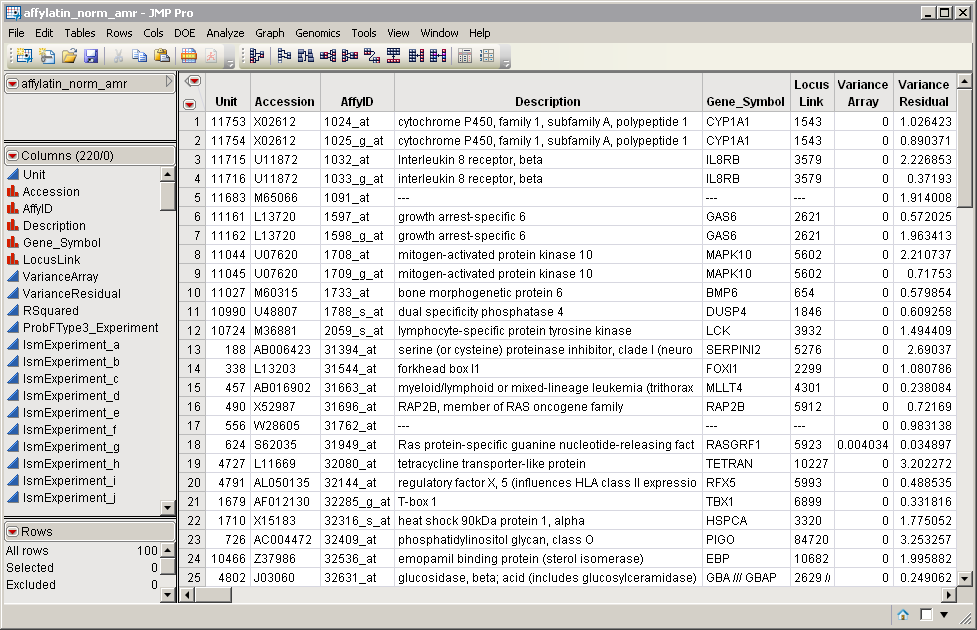
The affylatin_norm_amr.sas7bdat data set, located in the Sample Data\Microarray\Affymetrix Latin Square directory included with JMP Genomics, serves as an example, and is shown below.
This data set was generated from the Affymetrix Latin Square Data set, normalized, and subjected to ANOVA analysis. Genes or probesets are listed in rows, whereas annotation information and experimental summary statistics are listed in columns. The format of the gene identifiers, listed in the AffyID column, and the expression statistics, in subsequent columns, is supported by IPA.
Important: You must have either an Ingenuity Pathway Analysis System license or trial package to upload the output from this process and run the IPA analysis.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
Refer to the IPA Upload output documentation for detailed descriptions of the output of this process.