When SNP genotypes are imputed, genotypes are called with a degree of uncertainty. Imputed SNP-Trait Association tests for association between various types of traits and each individual SNP taking into account the probabilities for observing each possible genotype.
Two types of analyses can be performed: a general test based on the probabilities of each SNP genotype or a regression testing for a linear trend of SNP alleles. Adjustments can be made for quantitative covariates and random effects or for some trait types, strata variables. P-values from these tests, with adjustments applied if requested, are plotted along the marker map.
Please refer to the MIXED, GLIMMIX, LOGISTIC, and PHREG procedures in the SAS/STAT User's Guide for more information.
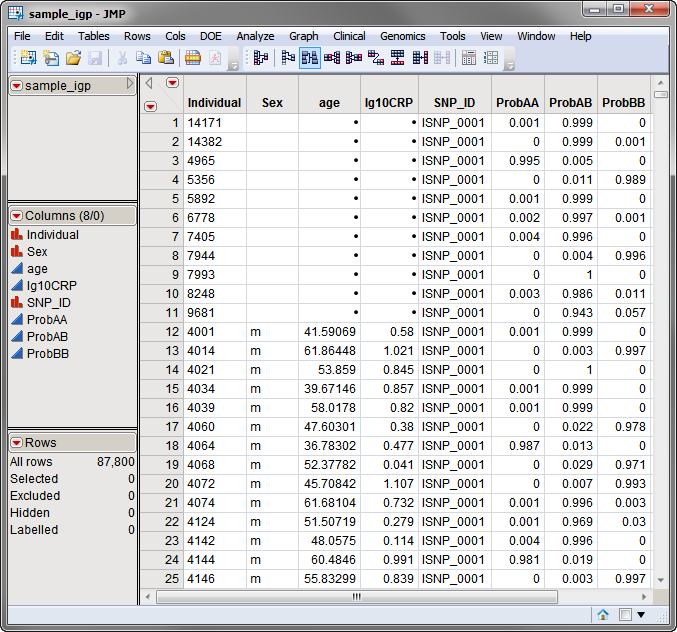
One Input Data Set, containing all of the marker data and genotypic probabilities, is required for this process. The Imputed SNP-Trait Association process differs from other Genetic Association processes in that the input data set used in this process must be in the stacked format, where there is a row for each SNP/individual combination, as created for example by the Imputed SNP input engines. The data set should be sorted by SNP.
The sample_igp.sas7bdat data set, shown below, was generated using the Imputed SNP (Wide Format) input engine, as described (Imputed SNP (Wide Format) Input Engine).
A second optional data set is the Annotation Data Set. This data set contains information, such as gene identity or chromosomal location, for each of the markers. This data set is a tall data set; each row corresponds to a different marker.
For detailed information about the files and data sets used or created by JMP Life Sciences software, see Files and Data Sets.
The output generated by this process is summarized in a Tabbed report. Refer to the Imputed SNP-Trait Association output documentation for detailed descriptions and guides to interpreting your results.