Suppose that your data consist of n rows and p columns. The rank of the covariance matrix is at most the smaller of n and p. In wide data sets, p is often much larger than n. In these cases, the inverse of the covariance matrix has at most n nonzero eigenvalues. Wide Linear methods use this fact, together with the singular value decomposition, to provide efficient calculations. See Calculating the SVD.
The singular value decomposition (SVD) enables you to express any linear transformation as a rotation, followed by a scaling, followed by another rotation. The SVD states that any n by p matrix X can be written as follows:
Diag(Λ) is an r by r diagonal matrix with positive diagonal elements given by the column vector 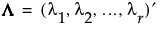 where
where 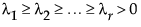 .
.
|
•
|
|
•
|
Note: There are various conventions in the literature regarding the dimensions of the matrices U, V, and the matrix containing the eigenvalues. However, the differences have no practical impact on the decomposition up to the rank of X.
Let n be the number of observations and p the number of variables involved in the multivariate analysis of interest. Denote the n by p matrix of data values by X.
The SVD is usually applied to standardized data. To standardize a value, subtract its mean and divide by its standard deviation. Denote the n by p matrix of standardized data values by Xs. Then the covariance matrix of the standardized data is the correlation matrix for X and is given as follows:
The SVD can be applied to Xs to obtain the eigenvectors and eigenvalues of Xs’Xs. This allows efficient calculation of eigenvectors and eigenvalues when the matrix X is either extremely wide (many columns) or tall (many rows). This technique is the basis for Wide PCA. See Wide Principal Components Options in Principal Components.
Denote the standardized data matrix by Xs and define S = Xs’Xs. The singular value decomposition allows you to write S as follows:
If S is not of full rank, then 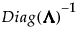 can be replaced with a generalized inverse,
can be replaced with a generalized inverse, 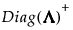 , where the diagonal elements of Diag(Λ) are replaced by their reciprocals. This defines a generalize inverse of S as follows:
, where the diagonal elements of Diag(Λ) are replaced by their reciprocals. This defines a generalize inverse of S as follows:
To see the specific details behind the application of the SVD for wide linear discriminant analysis, see Wide Linear Discriminant Method in Discriminant Analysis.
In the Multivariate Methods platforms, JMP’s calculation of the SVD of a matrix follows the method suggested in Golub and Kahan (1965). Golub and Kahan’s method involves a two-step procedure. The first step consists of reducing the matrix M to a bidiagonal matrix J. The second step consists of computing the singular values of J, which are the same as the singular values of the original matrix M. The columns of the matrix M are usually standardized in order to equalize the effect of the variables on the calculation. The Golub and Kahan method is computationally efficient.