Launch the Multivariate Embedding Platform
Launch the Multivariate Embedding Platform
Launch the Multivariate Embedding platform by selecting Analyze > Multivariate Methods > Multivariate Embedding.
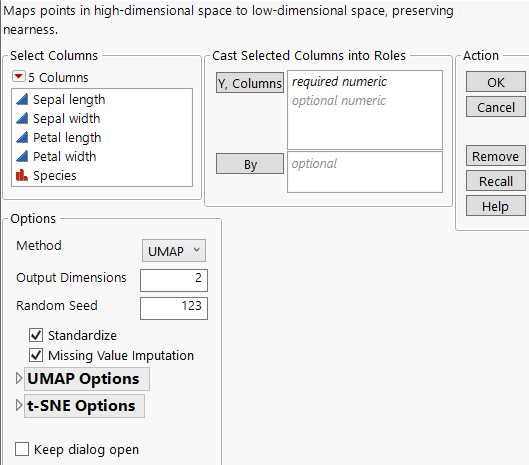
Figure 11.3 Multivariate Embedding Launch Window
For more information about the options in the Select Columns red triangle menu, see Column Filter Menu in Using JMP. The Multivariate Embedding launch window contains the following options:
Y, Columns
Specifies columns that represent the high-dimensional data to be mapped into a low-dimensional space.
By
A column whose levels define separate analyses. For each level of the specified column, the corresponding rows are analyzed using the other variables that you have specified. The results are presented in separate tables and reports. If more than one By variable is assigned, a separate analysis is produced for each possible combination of the levels of the By variables.
Method
Specifies the method for mapping the data into a low-dimensional space. Choose between UMAP and t-SNE.
Output Dimensions
Specifies the number of components or dimensions in the low-dimensional space. The number of components must be greater than or equal to 2.
Random Seed
Specifies a random seed to reproduce the results for future launches of the platform.
Standardize
Standardizes the data internally prior to computing the distances that are used in dimension reduction.
Missing Value Imputation
Specifies that missing values in the data are imputed using a multivariate singular value decomposition (SVD) technique.
Note: If your data contain missing values and you do not select the Missing Value Imputation option, an imputation window is shown once you click OK in the launch. If each row in the data contains at least one missing value, you can select to impute the missing values, change the selection of Y columns, or cancel the analysis. If some rows in the data do not contain missing values, you can select to impute the missing values, continue without imputation, or cancel the analysis.
UMAP Options
Contains options that are used in the UMAP algorithm. For more information on how the following parameters are used in the UMAP algorithm, see McInnes et al. (2018).
Number of Neighbors
Specifies the number of near neighbors that are found for each data point. The smaller the number of near neighbors specified, the more the UMAP algorithm concentrates on the local structure of the data. As the number of near neighbors increases, the UMAP algorithm captures more of the global structure of the data. The Number of Neighbors value can range from 2 to a quarter of the number of observations in the data. The default value is 15.
Number of Epochs
Specifies the number of training epochs to use when optimizing the low-dimensional representation. This is the number of times the algorithm works through the full training data. The default value is 500.
Learning Rate
Specifies the value of the learning rate in the computations. The default value is 1. The learning rate impacts how quickly the model adapts to the problem. If the learning rate is too large, the algorithm might miss the optimal solution. If the learning rate is too small, the algorithm might take a long time to converge.
Tip: If the algorithm does not converge or produces embedded coordinates that have extreme values, consider adjusting the value of the learning rate.
Minimum Distance
Specifies the minimum standardized distance that points in the low-dimensional space can be from one another. This value can range from 0 to 0.99. The default value is 0.01.
Local Connectivity
Specifies the number of nearest neighbors that are assumed to be connected at a local level. The default value is 1, which assumes that every point in the high-dimensional space has at least one other neighbor to which it is connected.
a
Specifies one of the parameters that control the embedding optimization algorithm. If this value is specified as 0 or a negative number, a is calculated in the algorithm by a nonlinear least squares procedure.
b
Specifies one of the parameters that control the embedding optimization algorithm. If this value is specified as 0 or a negative number, b is calculated in the algorithm by a nonlinear least squares procedure.
Negative Sample Rate
Specifies the number of negative 1-simplex samples to use per positive 1-simplex sample in finding the low-dimensional representation of the data. The Negative Sample Rate value can range from 2 to 20. The default value is 5.
Batch Mode if N Greater Than
Specifies that multithreading is used for optimizing the embedding coordinates when the sample size is larger than the specified number. The default value is 4096.
Nearest Neighbor Method
Specifies the method that is used for finding nearest neighbors.
Default
Chooses the nearest neighbor method according to the sample size and the number of variables. If the number of observations is greater than 4096 and the number of variables is less than or equal to 1500 or the Distance Metric is not set to Euclidean, the default is ANNOY. Otherwise, the default is VPTree.
VPTree (Exact)
Finds the set of nearest neighbors using a vantage-point (VP) tree.
ANNOY (Approximate)
Finds the set of nearest neighbors using the Approximate Nearest Neighbors (ANN) method (Bernhardsson, 2013). This is the faster of the two methods for large data sets, but the results might be less accurate than the VPTree method.
Distance Metric
(Applicable only when ANNOY is specified as the Nearest Neighbor Method.) Specifies the metric that is used to compute distances between nearest neighbors. The options for the distance metric are Euclidean, Angular, Hamming, and Manhattan. By default, Euclidean is specified as the Distance Metric.
Tip: If the data contain binary or categorical variables, a non-Euclidean distance metric might be more appropriate.
Gradient Descent Method
Specifies the gradient descent method that is used in the optimization algorithm.
SGD
Uses the Stochastic Gradient Descent algorithm (Saad, 1998). This is the default method.
ADAM
Uses the Adaptive Moment Estimation method (Kingma, 2014). This option is available only if multithreading is used.
t-SNE Options
Contains options that are used in the t-SNE algorithm. Many of these options are discussed in Statistical Details for the Multivariate Embedding Platform.
Sparse
Specifies whether sparse methods are used in the computation of the conditional probabilities in the high-dimensional space. Sparse methods enable computation for high-dimensional data.
Perplexity
Specifies the value of the perplexity parameter, which is related to computing similarities of the samples. The value of the perplexity parameter should be between 5 and 50 and should not be greater than one-eighth of the sample size. The default value is the smaller of 30 or one-eighth of the sample size.
Maximum Iterations
Specifies the maximum number of iterations that are used in the computations.
Initial Principal Component Dimensions
Specifies the number of dimensions that are retained in the initial random principal components analysis step. The default value is 50.
Convergence Criterion
Specifies the value that is used to measure convergence. The default value is 1e-8.
Initial Scale
Specifies the initial scale for the derived components. The default value is 0.0001.
Eta
Specifies the value of the learning rate in the computations. The default value is 200.
Inflate Iterations
Specifies the number of iterations after which the momentum value is no longer exaggerated. The default value is 250.
Keep dialog open
Keeps the launch window open after you run the analysis, enabling you to update options and re-run the analysis.