Publication date: 07/15/2025
Example of Distance Matrix
Learn how to compute various measures of distance or dissimilarity between observations in your life sciences genetic data.
1. Select Help > Sample Data Folder, and then open Iris.jmp.
2. Select Analyze > Life Sciences > Distance Matrix.
3. Select the first four columns (Sepal length, Sepal width, Petal length, and Petal width) and click Y, Columns.
4. Select Species and click X, Grouping Columns.
5. Click OK.
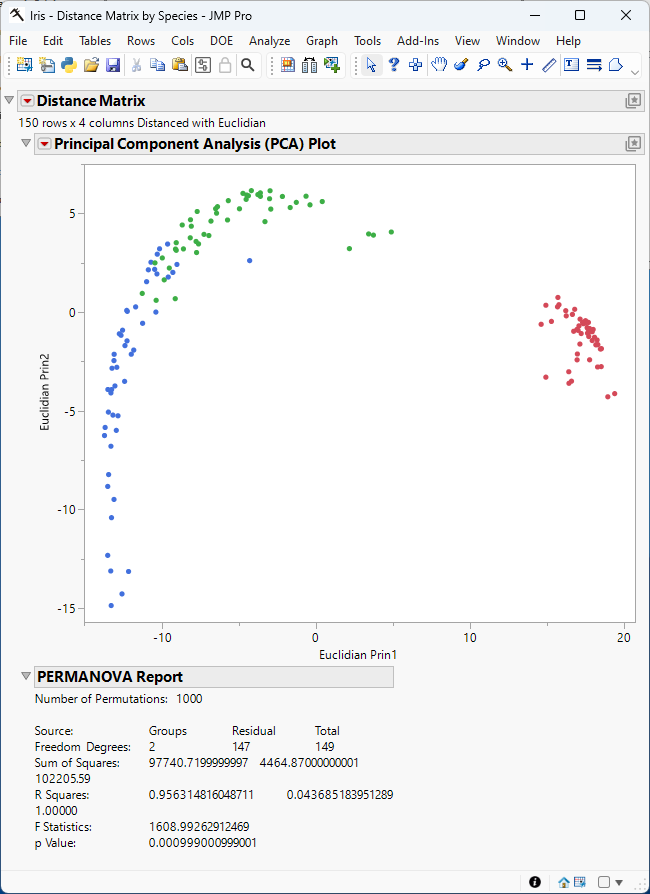
Figure 9.1 The Distance Matrix Report
The Principal Component Analysis (PCA) plot is shown above. Points corresponding to the observations in the input table are colored by species. The plot shows that the observations cluster by species and that variance in length and width of sepals and petals in irises is greatest between species.
Want more information? Have questions? Get answers in the JMP User Community (community.jmp.com).