Imputing Missing Values in Genotype Data Sets
Quite often, you can find that genomic data sets are found missing a large amount of data. This can be a real problem when you are trying to analyze your data. A number of the analysis platforms in JMP exclude rows with missing data, thus, much of your data could be lost from the analysis. In this case, you must find a way of imputing the missing data and filling the empty cells in your data table.
A number of multivariate platforms (Hierarchical Clustering and Principal Components, for example) have built-in imputation, which is a preferred method, when available. For other platforms, you can make an imputed version of your data using the Explore Missing Values platform.
In this example, we impute the missing data in the loblolly.jmp table of genotype and phenotype data described in Genetic Analysis of 6 Loblolly Pine Traits and shown below. This is a wide table containing 4890 columns and 926 rows.
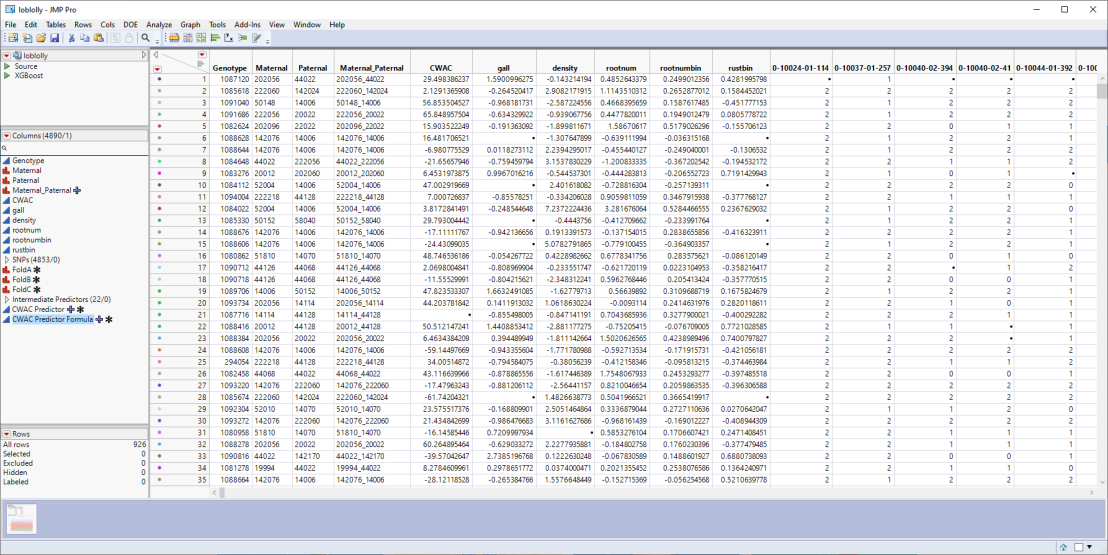
Missing data is indicated by a dot (.). This table is missing both genotype and phenotype data. We will use the Explore Missing Values platform to impute the missing values.
| 8 | Open the loblolly.jmp table. |
| 8 | Click Analyze > Screening > Explore Missing Values. |
| 8 | Select the columns to be imputed (in this case, all six trait columns and all of the SNP genotype columns) as the Y, Columns. |
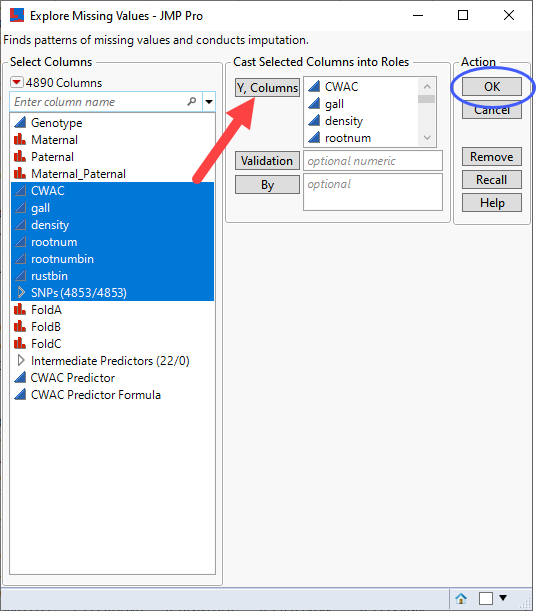
| 8 | Click . |
The following report is generated:
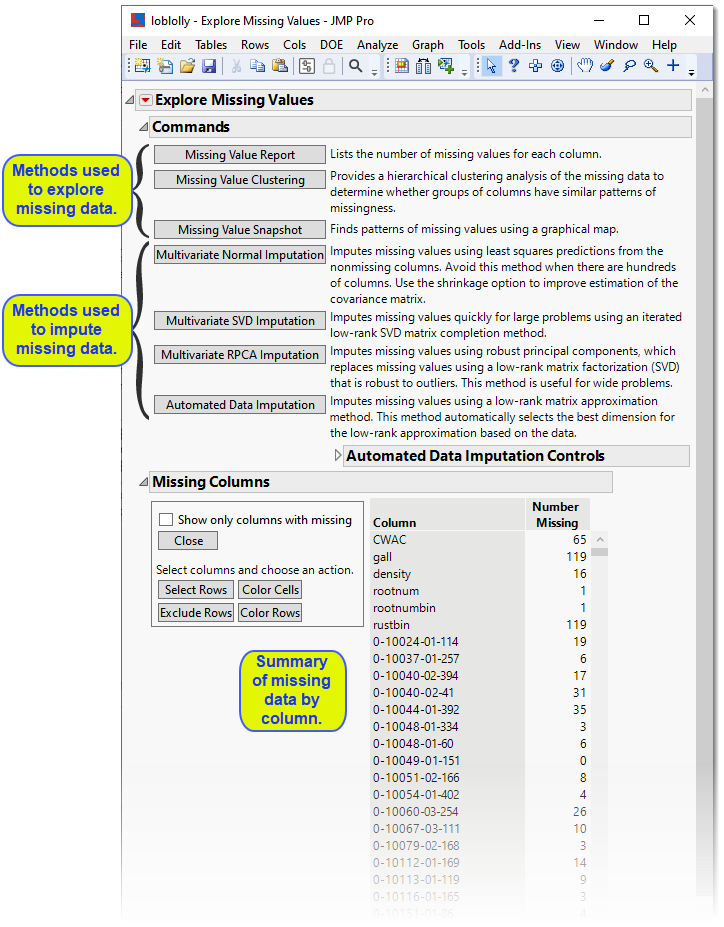
The report summarizes the missing data and provides numerous options for both exploring and imputing the missing data. You might want to experiment with imputation methods to find the best method for your data. In this example, we use .
| 8 | Click . |
The following data set is generated:
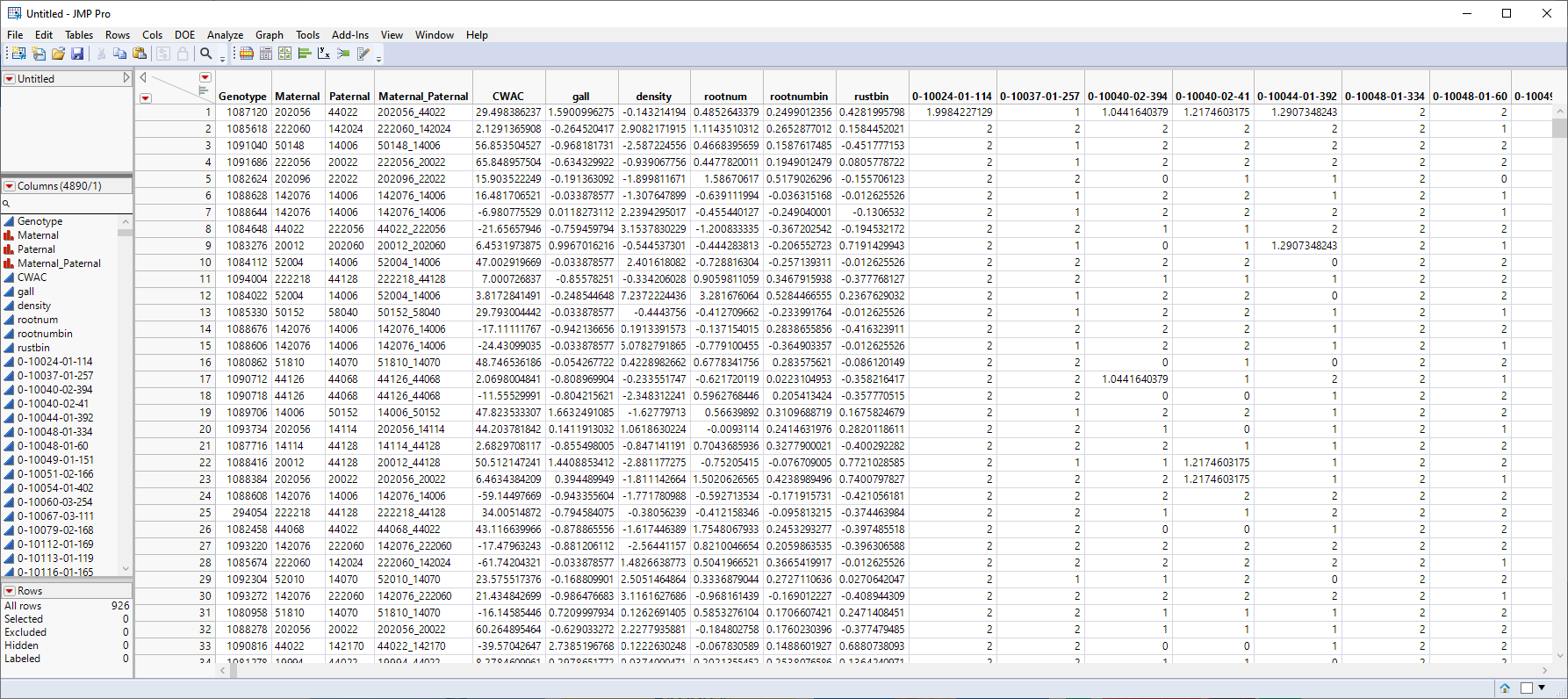
Note that the missing value symbols have been replaced with the imputed values.
| 8 | Save and name the new data set and proceed with your analysis. |