Verifying Marker Encoding
In this guide, we use a Wide table with markers in the columns and samples in the rows. This table was generated as described in the Importing VCF Files guide.
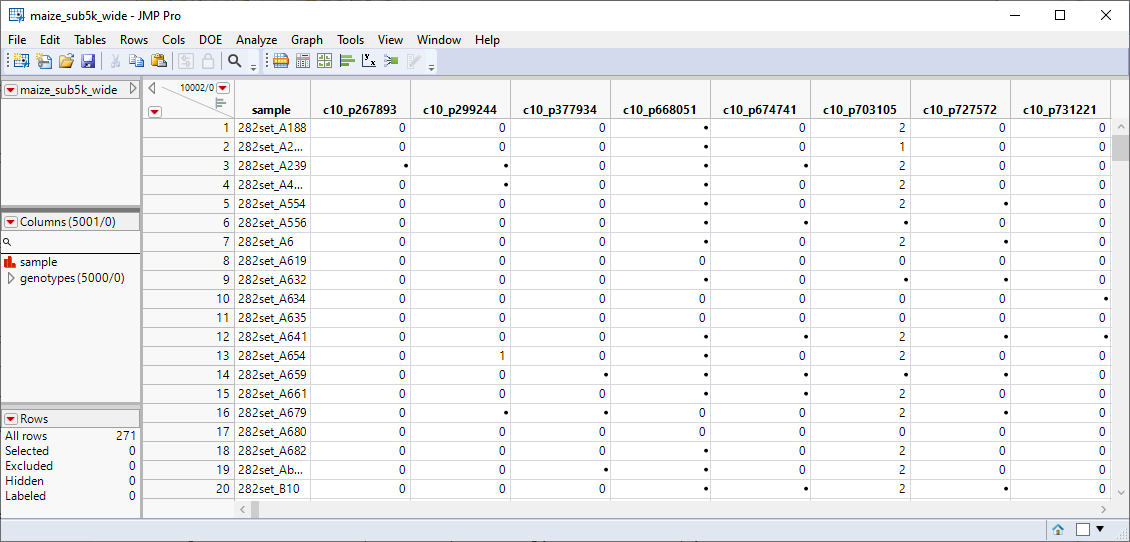
Before this data table is subjected to further analysis, you should ensure that all markers been coded in the same allelic count basis. For example, in a diploid species, individuals homozygous for the least common, or minor allele, are represented by a "2", whereas the heterozygotes are represented by a "1". Homozygotes for the most common allele are represented by a "0". It is perfectly reasonable to want the numeric code in the reverse order, that is,, 0 for the least common, 1 for heterozygous, and 2 for the most common. For polyploid species, is it also true that we need to make sure that all markers have been coded in the same allelic count basis.
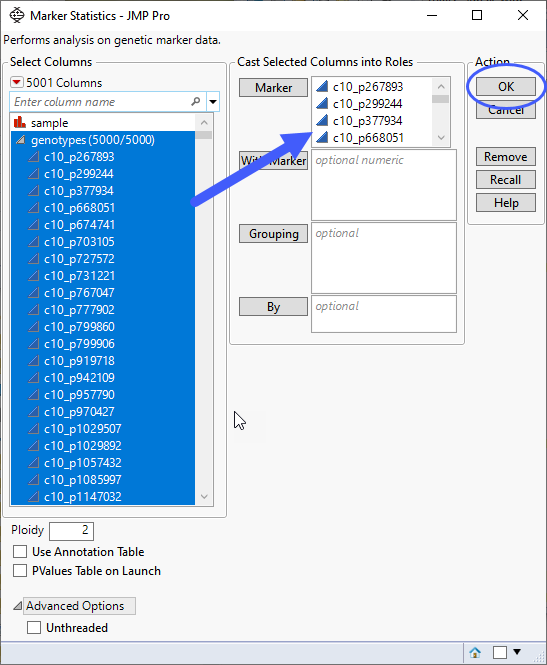
To make sure all markers have been numerically coded to the same allelic count basis, use the Marker Statistics platform (new in JMP Pro 17.0). This analysis consists of four steps:
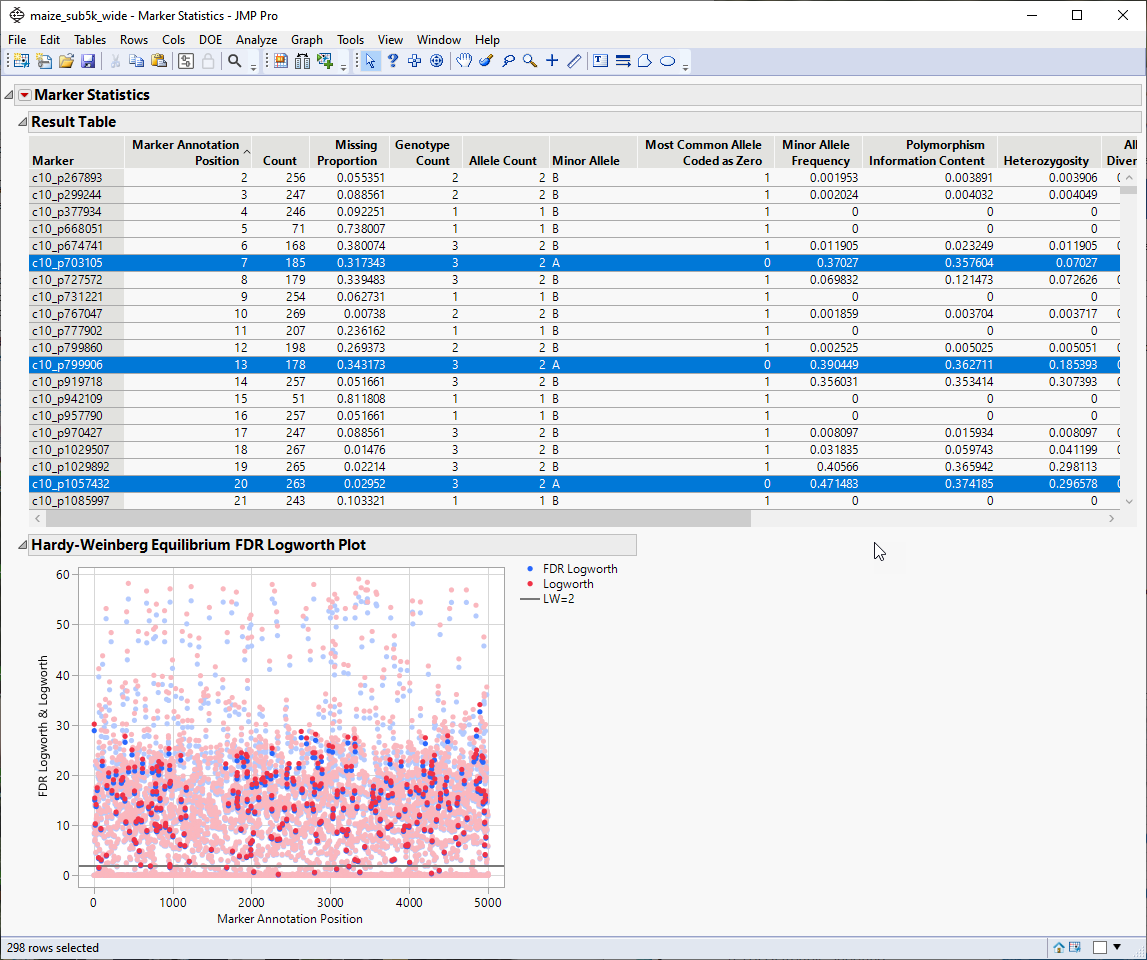
| 1 | Run Marker Statistics (Analyze > Genetics > Marker Statistics ) platform for all markers to obtain the Report window that contains the Results Table. |
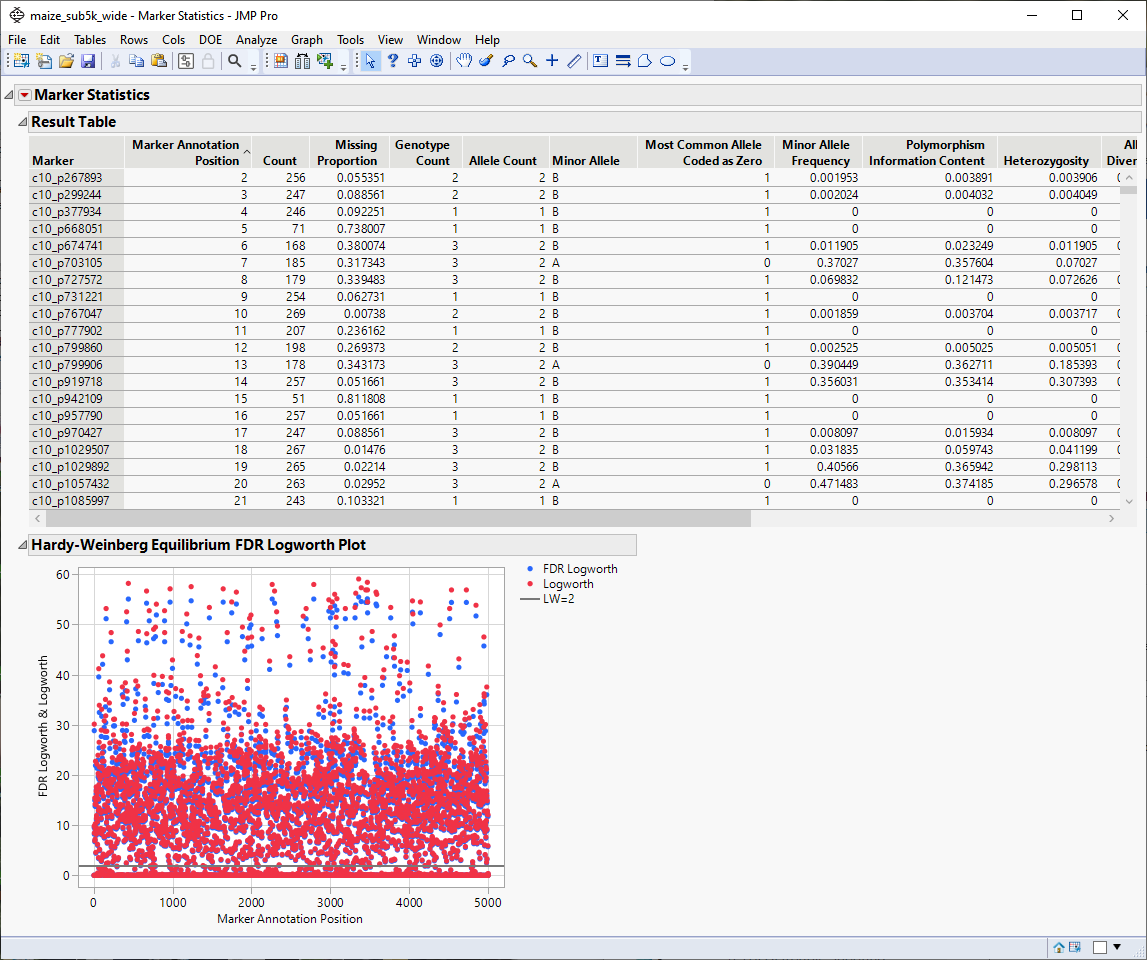
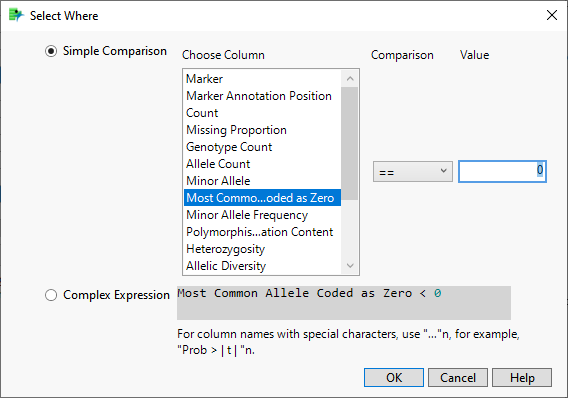
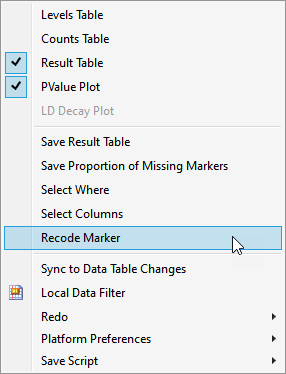
| 2 | In the Report Results window, run Select Where (from the red triangle ( ) menu) setting up Most Common Allele Coded as Zero == 0 (shown below). With this statement, only those markers for which the most common allele is not coded as "0" are selected in the Results Table. ) menu) setting up Most Common Allele Coded as Zero == 0 (shown below). With this statement, only those markers for which the most common allele is not coded as "0" are selected in the Results Table. |
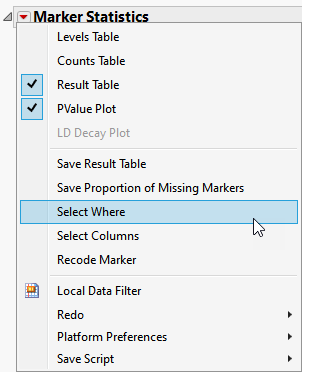
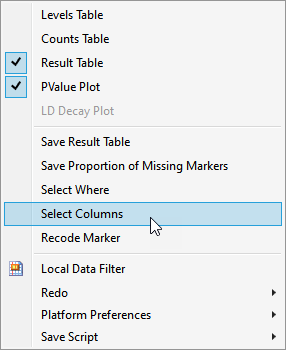
| 3 | In the Report results window, run Select Columns (in the red triangle) to select markers in the wide numeric data table that correspond to the selected rows in the Results Table. |
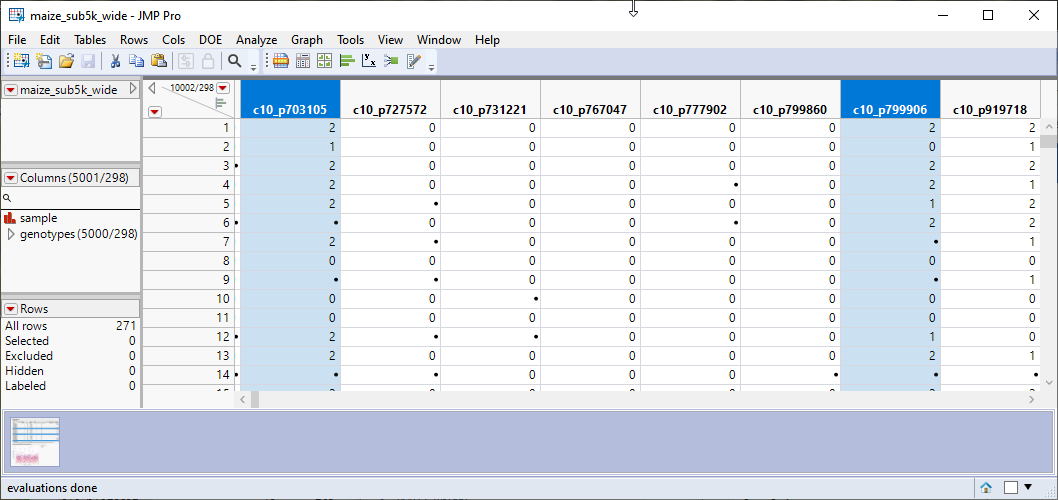
| 4 | In the Report results window, run Recode Marker (in the red triangle) to recode selected markers accordingly, that is,, 0→2, 1→1, 2→0. |
Note that the selected columns have been recoded.
When you obtain a new wide data table with numeric genotypes, it is highly recommended that you to run this data table through this procedure to make sure all markers have been coded using the same criteria.