Process Description
Import Feature-Barcode Matrices
This process imports 10x Genomics Single-Cell RNA Sequencing data to SAS data sets.
What do I Need?
You must specify one matrix (.mtx) file and two tab-separated value (.tsv) files.
The matrix file is a text file listing the matrix coordinates of the original counts files along with the counts at each location on the matrix. This file is specified in the Matrix File to Import field. For example, the matrix.mtx file, shown below, lists the x- and y- coordinates in the first two columns and the counts in the third.
The second required file is a text file listing the names of all the genes in the study. This file is specified in the Gene File to Import field. In the example shown below, the genes.tsv file has two columns listing alternate identifiers for each gene (rows).

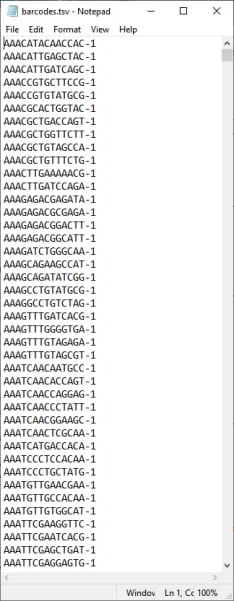
The third required file is a text file listing the identifiers (or bar codes) for each of the cells in the analysis. This file is specified in the Barcode File to Import field. In the example shown below, the barcodes.tsv file has one column listing the individual cell identifiers.
Note: The three files are merged to form the data set. The rows of each text file must match exactly.
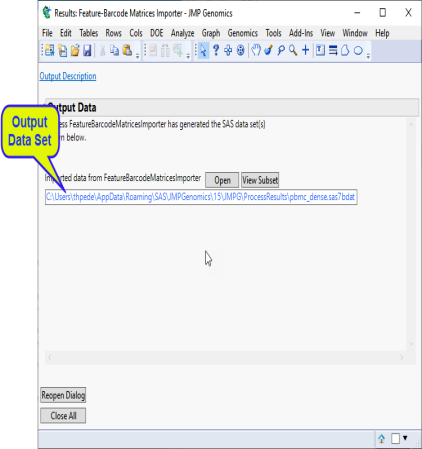
Output/Results
The output data sets generated by this process are listed in a Results window. Refer to the Import Feature-Barcode Matrices output documentation for detailed descriptions.