|
•
|
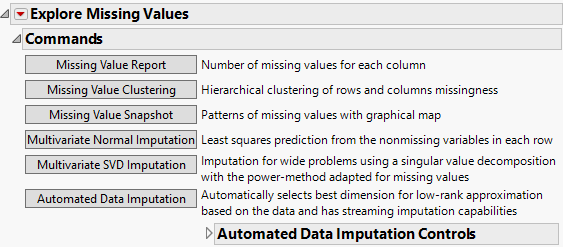
Multivariate Normal Imputation (Not available if you entered a Numeric column with a Nominal or Ordinal modeling type in the launch window.)
|
|
•
|
Multivariate SVD Imputation (Not available if you entered a Numeric column with a Nominal or Ordinal modeling type in the launch window.)
|
|
•
|
 Automated Data Imputation (Not available if you entered a Numeric column with a Nominal or Ordinal modeling type in the launch window.) Automated Data Imputation (Not available if you entered a Numeric column with a Nominal or Ordinal modeling type in the launch window.) |
Tip: To run a missing value command across all levels of a By variable, hold down the Ctrl key and click the desired command button.
Multivariate Normal Imputation allows the option to use a shrinkage estimator for the covariances. The use of shrinkage estimators is a way of improving the estimation of the covariance matrix. For more information about shrinkage estimators, see Schäfer and Strimmer (2005).
Click Undo to undo the imputation and replace the imputed data with missing values.
The singular value decomposition represents a matrix of observations X as X = UDV‘, where U and V are orthogonal matrices and D is a diagonal matrix.
The SVD algorithm used by default in the Multivariate SVD Imputation utility is the sparse Lanczos method, also known as the implicitly restarted Lanczos bidiagonalization method (IRLBA). See Baglama and Reichel (2005). The algorithm does the following:
|
3.
|
Each cell that had a missing value is replaced by the corresponding element of the UDV‘ matrix obtained from the SVD decomposition.
|
Click Undo to undo the imputation and replace the imputed data with missing values.
A low-rank approximation of a matrix is of the form X = UDV‘ and can be viewed as an extension of singular value decomposition (SVD). ADI uses the Soft-Impute method as the imputation model and is designed such that the data determines the rank of the low-rank approximation.
|
5.
|
|
6.
|
Figure 2.14 ADI Controls
Saves a column group, named Imputed_, to the current data table that contains the imputed columns specified in the launch window. A hidden column, ADI Impute Column, is also added to the current data table that contains the imputed vectors and the scoring formula used in the data imputation. The column formulas automatically update if any additional rows are added to the data table, enabling missing data imputation for streaming data. This is the default option.