Decompositions and Normalizations
This section contains functions that calculate eigenvalues and eigenvectors and functions that decompose matrices.
Eigenvalues
The Eigen() function performs eigenvalue decomposition of a symmetric matrix. Eigenvalue decompositions are used in many statistical techniques, most notably in principal components and canonical correlation, where the transformation associated with the largest eigenvalues are transformations that maximize variances.
Eigen() returns a list of matrices. The first matrix in the returned list is a column vector of eigenvalues; the second matrix contains eigenvectors as the columns.
A = [ 5 4 1 1, 4 5 1 1, 1 1 4 2, 1 1 2 4];
Eigen( A );
{[10, 5, 2, 1][0.632455532033676 - 0.316227766016838 - 2.77555756156289e-16 -0.707106781186547, 0.632455532033676 - 0.316227766016837 - 1.66533453693773e-16 0.707106781186547, 0.316227766016838 0.632455532033676 0.707106781186548 0, 0.316227766016837 0.632455532033676 - 0.707106781186547 0]}
Since the function returns a list of matrices, you might want to assign it to a list of two global variables. That way, the column vector of eigenvalues is assigned to one variable, and the matrix of eigenvectors is assigned to another variable:
{evals, evecs} = Eigen( A );
For some n × n matrix A, eigenvalue decomposition finds all λ (lambda) and vectors x, so that the equation Ax = λx has a nonzero solution x. The λ’s are called eigenvalues, and the corresponding x vectors are called eigenvectors. This is equivalent to solving (A - λI)x = 0. You can reconstruct A from eigenvalues and eigenvectors by a statement like the following:
newA = evecs * Diag( evals ) * evecs`;
[5.00000000000001 4 1 1,
4 5 1 1,
1 1 4 2,
1 1 2 4]
Note the following about eigenvalues and eigenvectors:
• The eigenvector matrices are orthonormal, such that the inverse is the transpose: 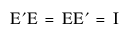 .
.
• Eigenvectors are uniquely determined only if the eigenvalues are unique.
• Zero eigenvalues correspond to singular matrices.
• Inverses can be obtained by inverting the eigenvalues and reconstituting with the eigenvectors. Moore-Penrose generalized inverses can be formed by inverting the nonzero eigenvalues and reconstituting. (See GInverse.)
Note: You must decide whether a very small eigenvalue is effectively zero.
• The eigenvalue decomposition enables you to view any square-matrix multiplication as the following:
– a rotation (multiplication by an orthonormal matrix)
– a scaling (multiplication by a diagonal matrix)
– a reverse rotation (multiplication by the orthonormal inverse, which is the transpose), or in notation:
A * x = E` * Diag( M ) * E * x;
E rotates, Diag( M ) scales, and E` reverse-rotates.
Cholesky Decomposition
The Cholesky() function performs Cholesky decomposition. A positive semi-definite matrix A is re-expressed as the product of a nonsingular, lower-triangular matrix L and its transpose: L*L' = A.
E = [ 5 4 1 1, 4 5 1 1, 1 1 4 2, 1 1 2 4];
L = Cholesky( E );
[2.23606797749979 0 0 0,
1.788854381999832 1.341640786499874 0 0,
0.447213595499958 0.149071198499986 1.9436506316151 0,
0.447213595499958 0.149071198499986 0.914659120760047 1.71498585142509]
To verify the results, enter the following:
L*L`;
[5 4 1 1,
4 5 1 1,
1 1 4 2,
1 1 2 4]
About Cholesky Decomposition
Cholesky() is useful for reconstituting expressions into a more manageable form. For example, eigenvalues are available only for symmetric matrices in JMP, so the eigenvalues of the product AB could not be obtained directly. (Although A and B can be symmetric, the product is not symmetric.) However, you can usually rephrase the problem in terms of eigenvalues of L′BL where L is the Cholesky root of A, which has the same eigenvalues.
Another use of Cholesky() is in reordering matrices within Trace() expressions. Expressions such as Trace(A*B*A`) might involve huge operations counts if A has many rows. However, if B is small and can be factored into LL′ by Cholesky, then it can be reformulated to Trace(A*L*L`*A`). The resulting matrix is equal to Trace(L`A`*AL). This expression involves a much smaller number of operations, because it consists of only the sum of squares of the AL elements.
Use the function Chol Update() to update a Cholesky decomposition. If L is the Cholesky root of a n × n matrix A, then after using cholUpdate(L, C, V), L will be replaced with the Cholesky root of A+V*C*V'. C is an m × m symmetric matrix and V is an n × m matrix.
Examples
Manually update the Cholesky decomposition, as follows:
exS = [16 1 0 11 -1 12, 1 11 -1 1 -1 1, 0 -1 12 -1 1 0, 11 1 -1 11 -1 9, -1 -1 1 -1 9 -1, 12 1 0 9 -1 12];
exAchol = Cholesky( exS ); // conducts the Cholesky decomposition
// add two column vectors to the design matrix
exV = [1 1, 0 0, 0 1, 0 0, 0 0, 0 1];
/* The first column vector is added to one of the rows in the design matrix.
The second column vector is subtracted from one of the rows in the design matrix. */
exC = [1 0, 0 -1];
exAnew = exS + exV * exC * exV`;
// update the Cholesky decomposition manually
exAcholnew = Cholesky( exAnew );
Instead of manual updating, use Chol Update() to update the Cholesky decomposition, as follows:
// update the Cholesky decomposition more efficiently
exAcholnew_test = Chol Update( exAchol, exV, exC );
Show( exAcholnew_test );
exAcholnew_test =
[ 4 0 0 0 0 0,
0.25 3.30718913883074 0 0 0 0, ...]
// results are the same as the manual process
Show( exAcholnew );
exAcholnew=
[ 4 0 0 0 0 0,
0.25 3.30718913883074 0 0 0 0, ...]
Singular Value Decomposition
The SVD() function finds the singular value decomposition of a matrix. That is, for a matrix A, SVD() returns a list of three matrices U, M, and V, so that U*diag(M)*V`=A.
Notes:
• When A is taller than it is wide, M is more compact, without extra zero diagonals.
• Singular value decomposition re-expresses A in the form USV′, where:
– U and V are matrices that contain orthogonal column vectors (perpendicular, statistically independent vectors)
– S is a n × n diagonal matrix containing the nonnegative square roots of the eigenvalues of A′A, the singular values of A.
• Singular value decomposition is the basis of correspondence analysis.
Example
A = [1 2 1 0, 2 3 0 1, 1 0 1 5, 0 1 5 1];
{U, M, V} = SVD( A );
newA = U * Diag( M ) * V`;
[1 2 0.999999999999997 -2.99456640040496e-15,
2 3 -1.17505831453979e-15 1,
0.999999999999997 -2.16851276518826e-15 0.999999999999999 5,
2.22586706011274e-15 1 5 0.999999999999997]
Orthonormalization
The Ortho() function orthogonalizes the columns and then divides the vectors by their magnitudes to normalize them. This function uses the Gram-Schmidt method. The column vectors of orthogonal matrices are unit-length and are mutually perpendicular (their dot products are zero).
B = Ortho( [1 -1, 1 0, 0 1] );
[0.408248290463863 -0.707106781186548, 0.408248290463863 0.707106781186548, -0.816496580927726 3.14018491736755e-16]
To verify that these vectors are orthogonal, multiply B by its transpose, which should yield the identity matrix.
C = B` * B;
[1 -3.119061760824e-16, -3.119061760824e-16 1]
By default, vectors are normalized, meaning that they are divided by their magnitudes, which scales them to have length 1. Include the option Scaled(0) to turn scaling off:
Ortho( [1 -1, 1 0, 0 1], Scaled( 0 ) );
[0.408248290463863 -0.353553390593274, 0.408248290463863 0.353553390593274, -0.816496580927726 1.57009245868377e-16]
To create vectors whose elements sum to zero, include the Centered(1) option. This option is useful when constructing a matrix of contrasts.
result = Ortho( [1 -1, 1 0, 0 1], Centered( 1 ) );
[0.408248290463863 -0.707106781186548, 0.408248290463863 0.707106781186548, -0.816496580927726 3.14018491736755e-16]
To verify that the elements of each column sum to zero, premultiply by a vector of ones to sum the columns.
J(1, 3) * result;
[1.11022302462516e-16 2.02996189274239e-16]
Orthogonal Polynomials
The Ortho Poly() function returns orthogonal polynomials for a vector of indices. The polynomial order is specified as a function argument. Orthogonal polynomials can be useful for fitting polynomial models where some regression coefficients might be highly correlated.
Ortho Poly( [1 2 3], 2 );
[-0.707106781186548 0.408248290463862, 0 -0.816496580927726, 0.707106781186548 0.408248290463864]
The polynomial order must be less than the dimension of the vector. Use the Scaled(1) option to produce vectors of unit length, as described in Orthonormalization.
QR Decomposition
QR() factorization is useful for numerically stable matrix work. QR() returns a list of two matrices. Here is an example:
{Q, R} = QR( [11 22, 33 44] );
Q and R hold the results. For a m × n matrix, QR() creates an orthogonal m × m matrix Q and an upper triangular m × n matrix R, so that X = Q*R.
Update Inverse Matrices
To add or drop one or more rows in an inverse of an M`M matrix, use the Inv Update(S, X, w) function. Updating inverse matrices is helpful in drop-1 influence diagnostics and also in candidate design evaluation.
Notes:
• The first argument, S, is the matrix to be updated.
• The second argument, X, is the matrix of rows to be added or dropped.
• The third argument, w, is either 1 to add or -1 to delete the row or rows.
• Multiple rows can be added or deleted.
Using the Inv Update(S, X, w) function is equivalent to calculating the following:
S - w * S * X` * Inv( I + w * X * S * X` ) * X * S
Where I is an identity matrix and Inv(A) is an inverse matrix of A.