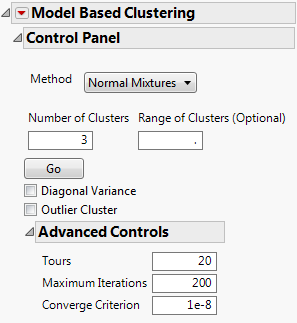
Model Based Clustering Control Panel
The Control Panel for the Cytometry.jmp data table, with the variables CD3 through MCB as Y, Columns, is shown in Figure 14.5. You can fit various numbers of clusters using the Control Panel iteratively or you can specify a range using the Range of Clusters option.
Figure 14.5 Control Panel for Normal Mixtures Method
The Model Based Clustering Control Panel has these options:
Number of Clusters
Designates the number of clusters to form.
Range of Clusters (Optional)
Provides an upper bound for the number of clusters to form. If a number is entered here, the platform creates separate analyses for every integer between Number of clusters and the value entered as Range of Clusters (Optional).
Go
Fits the clusters.
Diagonal Variance
Constrains the off-diagonal elements of the covariance matrix to zero. The platform fits multivariate normal distributions that have no correlations between the variables.
Note: The Diagonal Variance option is sometimes necessary to avoid obtaining a singular covariance matrix when there are fewer observations than variables. It can also be used to avoid estimating very large covariance matrices for large numbers of variables.
Outlier Cluster
Fits a cluster to catch outliers that do not fall into any of the normal clusters. If this cluster is created, it is designated Cluster 0, and the count of observations appears in the Cluster Summary report. The distribution of observations that fall in the outlier cluster is assumed to be uniform over the hypercube that encompasses the observations.
Advanced Controls
The following advanced controls are available:
Tours
The number of independent restarts of the estimation process. Each restart has a different starting value. Independent starts help guard against finding local solutions.
Maximum Iterations
The maximum number of iterations of the convergence stage of the EM algorithm.
Converge Criterion
The difference in the likelihood at which the EM iterations terminate.