Example of Marker Imputation
Learn how to impute missing marker data in a JMP table.
1. Select Help > Sample Data Folder, open Life Sciences, and then open Genotypes Pedigree.jmp.
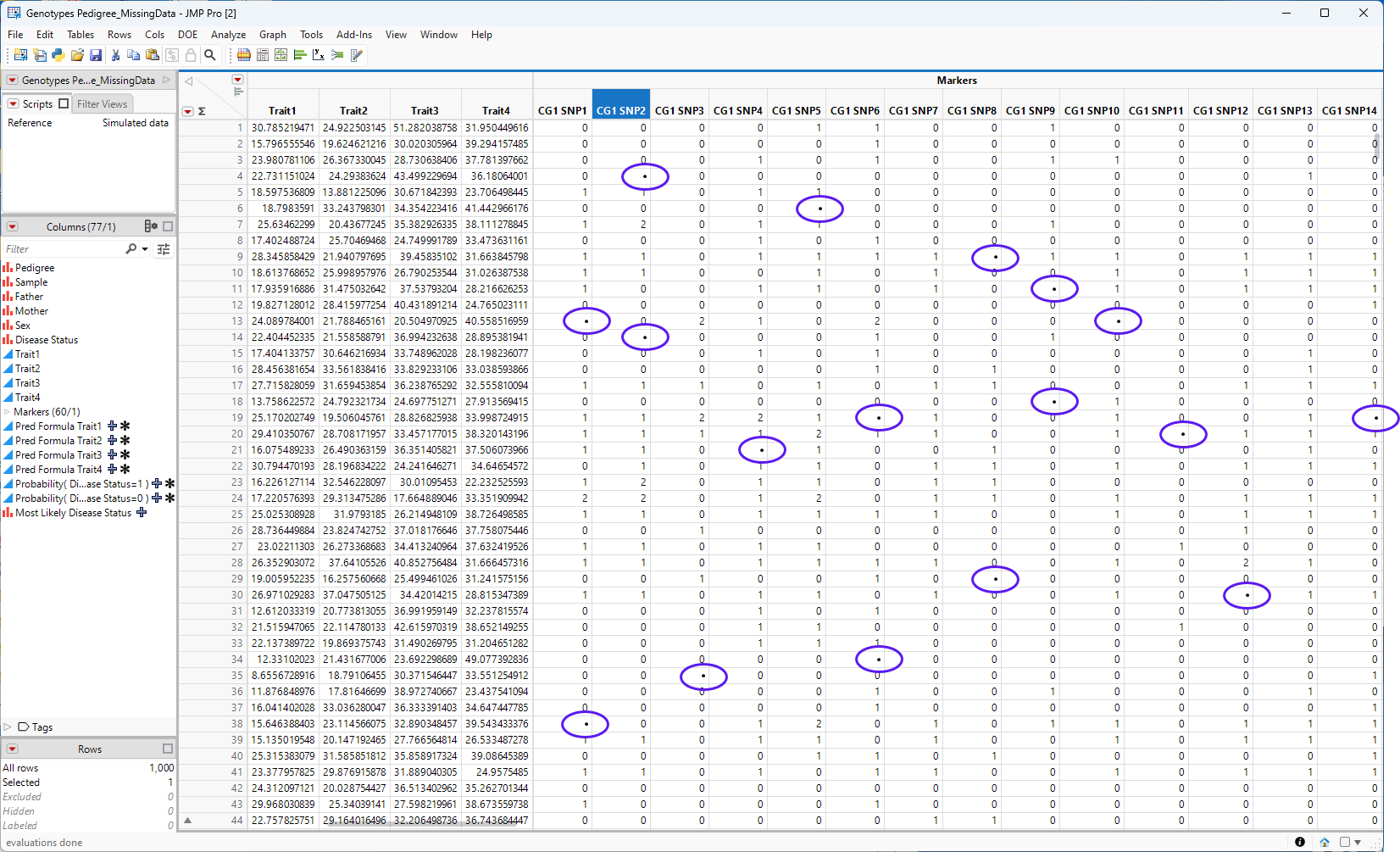
The Genotypes Pedigree.jmp table contains no missing data, so for this example, we had to manufacture some 17 cells in the first 14 columns of the marker data (columns CG1SNP1 through CG1SMP30) were selected at random and the data within each cell was erased. The resulting cells all contained a missing data symbol (•) This table was saved as the GENOTYPES pedigree_MissingData.jmp table, shown below. The cells all containing missing data now contain a missing data symbol (•)(circled in **).
Figure 7.2 The Genotypes Pedigree_MissingData.jmp JMP table containing missing data
2. Select Analyze > Life Sciences > Marker Imputation.
3. Select the Markers and click Marker.
4. Click OK.
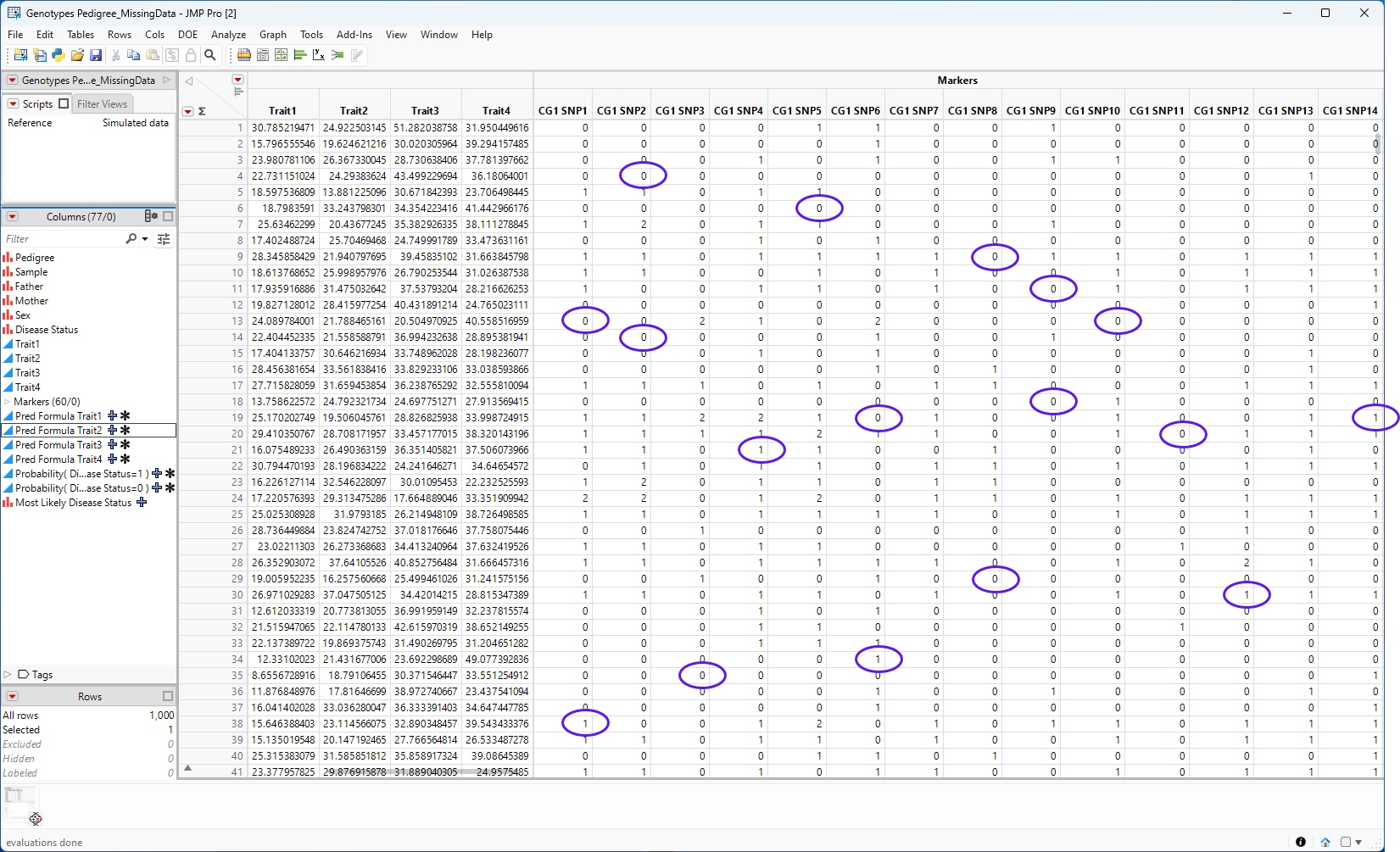
The platform assesses the missing data and, depending on the imputation method chosen, imputes the missing genotype data and adds the imputed genotypes to the empty spaces in the table, as shown below.
Figure 7.3 The completed table
The imputed genotypes in the completed table are circled.