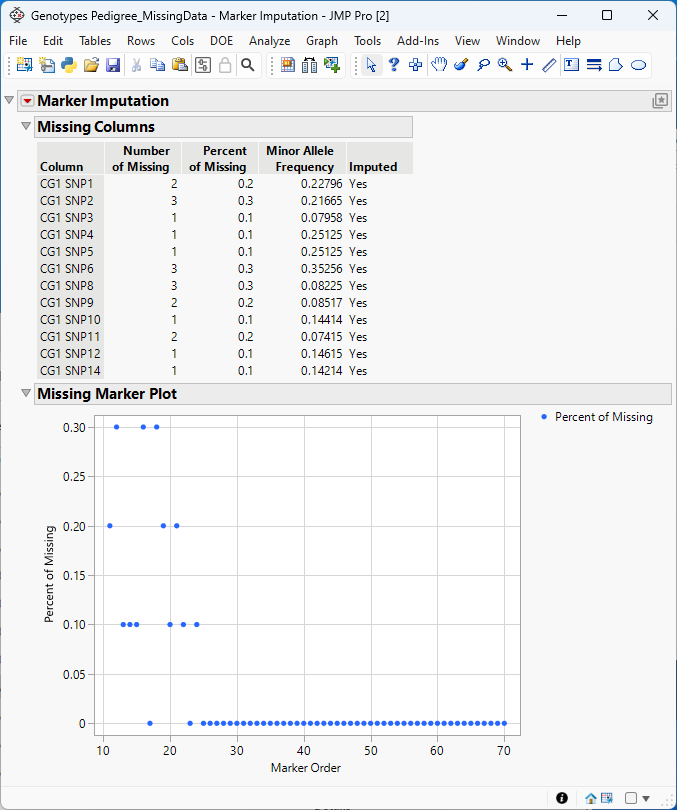
Marker Imputation
The Marker Imputation platform imputes numeric missing marker genotypes (0, 1, ..., K) for diploid (where K=2) or polyploid (K>2) organisms using the linkage disequilibrium k-nearest neighbor imputation (LD-kNNi) imputation or other methods. Distances (Pearson's correlation) between markers with missing genotypes and all other markers are computed and for each marker with a missing genotype, a set of closest markers is selected. Next, the respective selected set of markers is used to compute distances (taxi cab distance) between samples with missing genotypes and all other samples. Finally, the set of closest samples and closest markers are used to impute the missing genotypes (see Money et al., 2015).
Note: The Marker Imputation platform is available only in JMP Pro.
Figure 7.1 Example analysis of missing data in a Table containing genetic marker data