Launch the Normalization Platform
Launch the Distance matrix platform by selecting Analyze > Life Sciences . Distance Matrix.
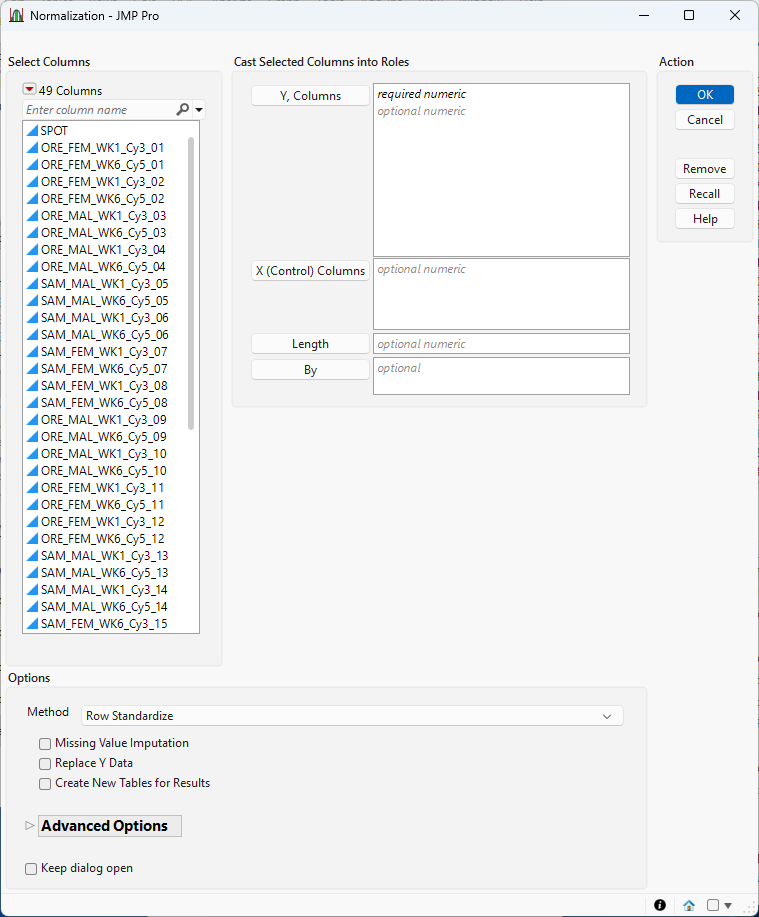
Figure 8.2 The Normalization Launch Window
For information about the options in the Select Columns red triangle, see “Column Filter Menu”in Using JMP.
Y, Columns
Select the marker columns that you want and click Marker to specify the markers that you want to analyze.
X, Control Columns
.Use this option to specify known control or reference columns, such as spike-ins or housekeeping genes, that are expected to stay stable across all samples. These helps anchor normalization when using methods like TMM.
Length
Use this option to specify the column containing the length of each transcript.
By
Produces a separate report for each level of the By variable. If more than one By variable is assigned, a separate report is produced for each possible combination of the levels of the By variables.
Method
Select the algorithm to use for performing the normalization.
• Select Row Standardize to scale each row to have a mean of 0 and standard deviation of 1. This method requires continuous data; it is not ideal for compositional data or raw counts. This method is useful for highlighting relative variation within samples, especially in conjunction with clustering and principal components analysis.
• Select Relative Abundance Percentage (RABP) to convert raw counts into percentage abundances across each sample. This method is used for counts data. It is used to emphasize relative proportions; it should not be used for statistical models that are sensitive to compositional bias. It is particularly useful for visualizations that emphasize relative amounts, such as pie charts or bar graphs and for exploratory analysis.
• Select Relative Abundance Ratio (RAR) to expresses each feature's abundance as a ratio relative to a reference (e.g., a control or geometric mean). This method is used for counts data. It is used to highlight fold changes relative to a baseline. It is not ideal for either skewed or sparse data.
• Select Centered Log Ratio (CLR) to log-transform compositional data centered on the geometric mean. This method is used for removing the closure effect in compositional data. It is particularly suited for microbiome data and metagenomics. It is best not to use this method with data that contains ”zero” counts.
• Select Count/Read Per Million (CPM/RPM) to scale counts by total reads in each sample and then multiply by one million. This method is used to adjust sequencing data to account for differences in sequencing depth when investigating expression comparisons across samples. It is best not to use this method with data that contains ”zero” counts.
• Select Read/Fragment Per Kilobase of Transcript Per Million Mapped Reads/Fragments (RPKM)?FPKM) to normalize sequencing data for both gene length and sequencing depth. This method is used for counts data. It is particularly useful with transcriptonic expression profiling and within-sample comparisons. Because gene length is not considered, it is best not to use this method for between sample comparisons.
• Select Transcript Per Million (TPM) to normalize sequencing data for both gene length and sequencing depth. This method is similar to RPKM, but more suitable for between-sample comparison. This method is used for counts data. Reduces compositional bias in RNA-seq. It is best for: differential expression (e.g., edgeR) data, but should not be used for very sparse or low-depth datasets.
• Select Trimmed Mean of M-Values (TMM) to perform a normal or log-ratio normalization using a reference sample, trimmed to reduce bias. This method reduces compositional bias in RNA-seq and is best for differential expression (e.g., edgeR) data. It is not ideal for very sparse or heterogeneous datasets.
• Select Kernel Density Mean of M-Component (KDMM) to perform a normal or log-ratio density-aware normalization using smoothed M-component distributions. This method takes weighted means of log-ratios across kernel-estimated density regions and adjusts for multimodal and nonlinear compositional structure. It is used on counts or compositional data. It is best for advanced microbiome and metagenomics normalization but should not be used for small sample sizes or low-resolution data.
Missing Value Imputation
Check this option when there are missing values to be imputed.
Replace Y-Data
Check this option to replace the original Y-columns with the corresponding normalized Y-columns.
Create New Tables for Results
Check this option to create new tables for the output data and results. When this option is unchecked output is added to the end of the input data table.
M Threshold
Use this advanced option to set a cutoff for log fold-change (M-values) between samples. This trims out extreme values that might distort normalization. Formula: M = log2(sample / reference). Use of this option helps improve stability, especially in datasets with strong differential expression.
This option is used for TMM and KDMM only.
A Threshold
Use this advanced option to apply a lower limit to average log-intensity (A-values) so that low-abundance or noisy features don't skew the normalization. Formula: A = 0.5 × log2(sample × reference)
This option is used for TMM and KDMM only.
Prior Count
Use this advanced option to add a small constant to all counts before log transformation (e.g., log2(count + 0.5)). This prevents issues with zeros in sparse datasets.
This option is used for TMM and KDMM only.
Interpolated Quantiles
Use this advanced option to enable the use of smoothed quantiles during normalization, which can be especially helpful in KDMM for complex or uneven data distributions
This option is used for TMM and KDMM only.
Log Transformed
Use this advanced option to convert the raw counts to log-transformed values.
This option is used for TMM and KDMM only.
Keep dialog open
Check this box to keep this platform dialog open after the analysis is run.
Data Format
Most of the processes in JMP assume that the input table has a particular data structure. JMP distinguishes between tall and wide data sets. A tall data table has samples as columns and molecular entity (for example, marker, gene, clone, protein, or metabolite) as rows, whereas a wide data table is the transpose of the tall data table, having the samples as rows and molecular entity as columns.
When specifying the input data set for a process, it is important to know the required form. Marker Imputation requires a wide data table. The Transpose platform under the Tables menu enables you to transform your data tables between tall and wide forms.
Marker data must be encoded in the one-column, numeric format. Typically, in this format, diploid individuals homozygous for the least common, or minor allele, are represented in the table by a 2, whereas the heterozygotes are represented by a 1. Homozygotes for the most common allele are represented by a 0.