Additional Examples of the Functional Data Explorer Platform
Additional Examples of the Functional Data Explorer Platform
• Example for Multiple Functional Processes
 Example for Multiple Functional Processes
Example for Multiple Functional Processes
This example uses the Fermentation Process.jmp and Fermentation Process Batch Yield Results.jmp sample data tables to analyze enzyme production. Yield is the amount of an enzyme produced by genetically modified yeast. There are 100 process measurements per batch that were taken at equally spaced times over a 12-hour period.
Use the Functional Data Explorer platform to fit models to the data and save functional principal components to a new data table. The functional principal components are then analyzed in the Generalized Regression personality of the Fit Model platform.
Fit Functional Models
1. Select Help > Sample Data Library and open Functional Data/Fermentation Process.jmp.
2. Select Analyze > Specialized Modeling > Functional Data Explorer.
3. In the Stacked Data Format tab, select Ethanol through pH and click Y, Output.
4. Select Time and click X, Input.
5. Select BatchID and click ID, Function.
6. Click OK.
7. Click the Functional Data Explorer Group red triangle and select Data Processing > Align > Align Range 0 to 1. This aligns the input variable to be between 0 and 1 in each Functional Data Explorer report.
8. Click the Functional Data Explorer Group red triangle and select Models > B-Splines. This fits a B-spline model to each of the functional processes.
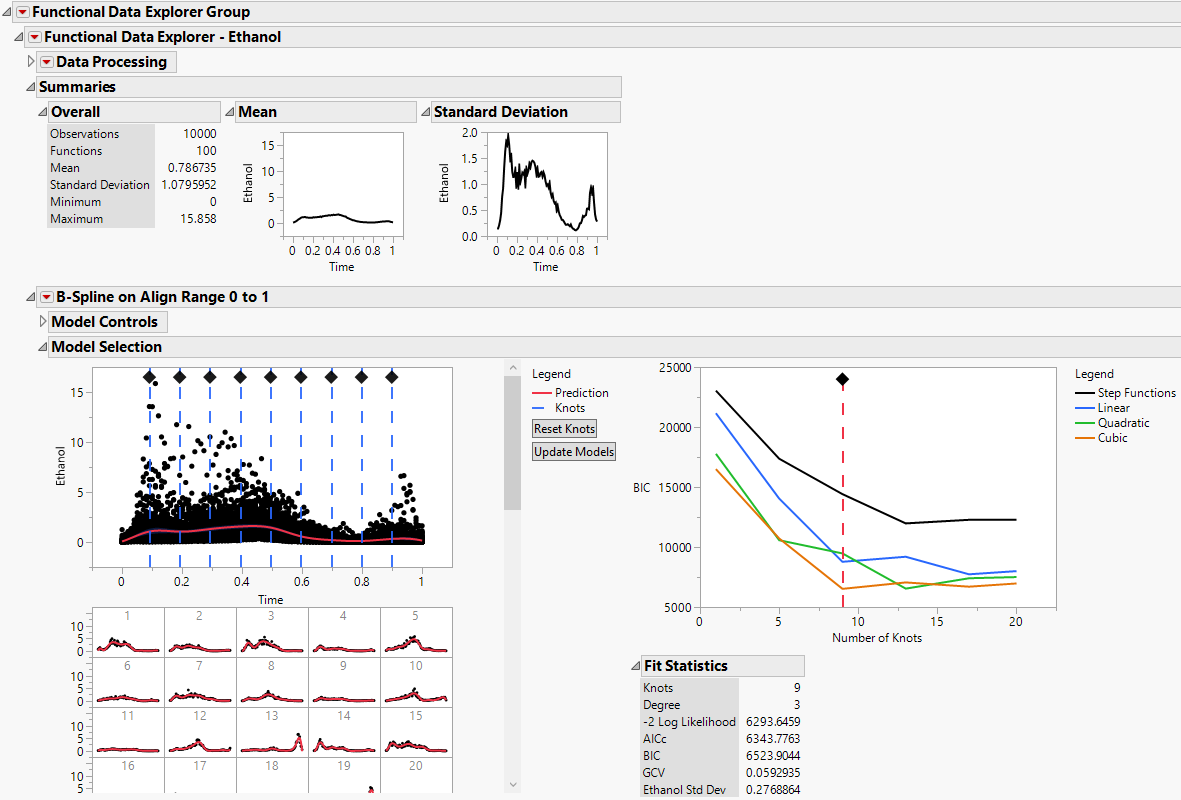
Figure 15.7 Functional Data Explorer Report for Ethanol
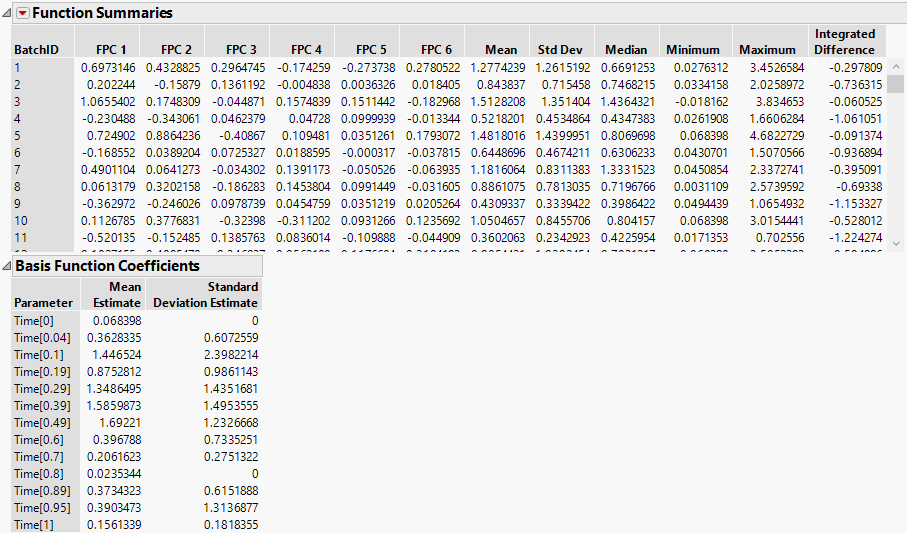
Figure 15.8 Model Summary Report for Ethanol
Figure 15.7 and Figure 15.8 show the model reports for one of the functional process variables, Ethanol. Scroll through the full report to view the models fit for each of the process variables. Next, use the FPCs in the Function Summaries report in an analysis.
Save FPCs and Link Yield Results
1. Press Ctrl, click any Function Summaries red triangle, and select Customize Function Summaries.
2. In the box next to Enter number of FPCs to show, type 3.
3. Click the Deselect All Summaries box.
4. Click OK.
5. Click the Functional Data Explorer Group red triangle and select Save Summaries.
6. Select Help > Sample Data Library and open Functional Data/Fermentation Process Batch Yield Results.jmp.
7. In the Functional Data Explorer Model Summaries.jmp data table, right-click BatchID and deselect Link ID.
8. In the Functional Data Explorer Model Summaries.jmp data table, right-click BatchID and select Link Reference > Fermentation Process Batch Yield Results.jmp.
This virtually joins the yield data table and the summaries data table.
Fit a Generalized Regression Model
Use the Generalized Regression personality of the Fit Model platform to determine how Yield is affected by the functional process variables.
1. In the Functional Data Explorer Model Summaries.jmp data table, select Analyze > Fit Model.
2. Click the triangle next to referenced by BatchID to Fermentation Process Batch Yield Results.
3. Select Yield[BatchID] and click Y.
4. Select the remaining columns, except Time and BatchID, and click Add.
5. Change the Personality to Generalized Regression.
6. Click Run.
7. Select the Adaptive box.
8. Click Go.
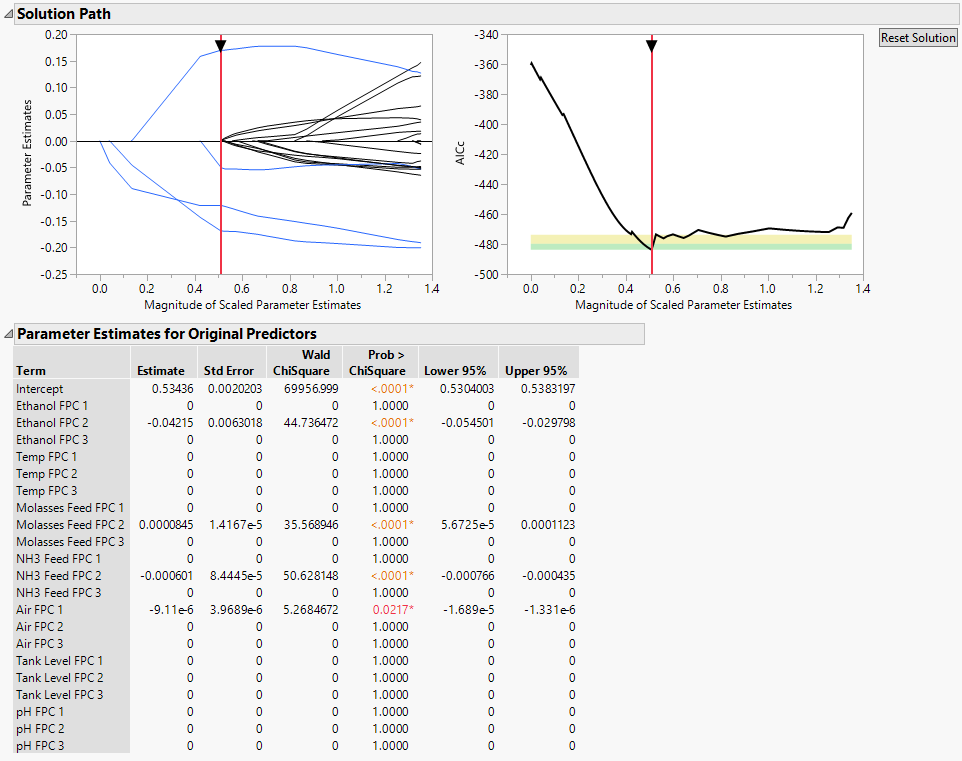
Figure 15.9 Generalized Regression Report for Batch Yield
The Generalized Regression report shows that Yield is significantly affected by certain components of Ethanol, Molasses Feed, NH3 Feed, and Air. The RSquare for the model is 0.73225. By using FDE to perform dimension reduction on the functional processes first, you greatly reduce the number of variables, while still retaining the ability to build a reasonable prediction models.