Cluster Subjects across Study Sites
This report is used to identify similar subjects. It does so by constructing a cross domain data set using as much data as possible (subject to user options). Next, it calculates Euclidean distances to compute a distance matrix and performs hierarchical clustering of subjects, across all of the study centers. Findings values are averaged by USUBJID, test code, visit number, and time point (if available) if there are multiple measurements for a visit or time point. The goal of this exercise is to identify pairs of subjects with a very small distance. This could be an indication that these subjects are in fact the same individual who has enrolled at multiple sites.
Report Results Description
Running this report using the Nicardipine sample setting and default options generates the output shown below. This report uses pre-dosing information with the goal of identifying subjects that have enrolled at two or more clinical sites.
The Cluster Subjects Across Study Sites report shows the results of clustering of the subjects on the basis of different combinations of covariates (demographic groups in this example). The results for each grouping are presented on a separate “section”. This report initially shows two sections Between-Subject Distance Summary and Subgroup Clustering. Use the available options in each section to drill-down into the data.
Results Sections
This pane enables you to access and view the output plots and associated data sets on each section. Use the drop-down menu to view the section in the Results pane or remove the section and its contents from the Results pane.
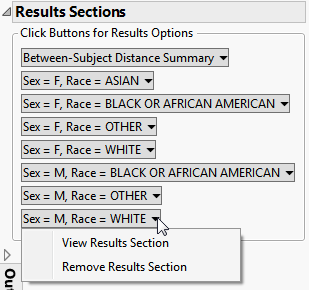
Between-Subject Distance Summary
| • | Box plots are presented for all pairwise distances between subjects in the selected population. Pairs are limited based on selections from the Cluster subjects matching these criteria panel of the dialog. |
In this example, box plots are presented by gender and race. The more similar a pair of subjects, the smaller the distance value (a zero indicates a perfect match).
| • | One Box Plot of Minimum Between-Subject Distances for Each Site. The minimum distance from each covariate subgroup is presented in the box plot to the right. |
The subgroup with the most similar pair of subjects is presented in the Subgroup Clustering section.
| • | A Local Data Filter to subset histograms to data of interest is available. |
In this example, data are filtered for a particular set of study monitors. Age (and height and weight, if available) are presented to limit pairs to those that are more likely to indicate a match between two subjects. Selecting Within subsets to pairs from subjects within the same site. Selecting Between subsets to subjects from different sites.
The data filters for this report include an option to specify the Number of Overlapping Variables. One of the challenges in this report is that distances may be very small, even zero. This may be driven by the fact that a patient may have a number of missing values for variables, and these don't contribute to the distance calculation. By default, SAS calculates the distance between pairs of variables that are non-missing for each pair of subjects. Number of Overlapping Variables makes it easier to subset to pairs that have a high-number of non-missing overlapping pairs of variables.
Refer to the Data Filter documentation for more information
Subgroup Clustering
One or more Subgroup Clustering sections: Only one section is opened initially. The name of this section is dependent on the covariate values used (as specified in the Cluster subjects matching these criteria panel) and the subgroup that is identified with the minimum pairwise distance. Other subgroup results can be opened from the Results Sections menu.
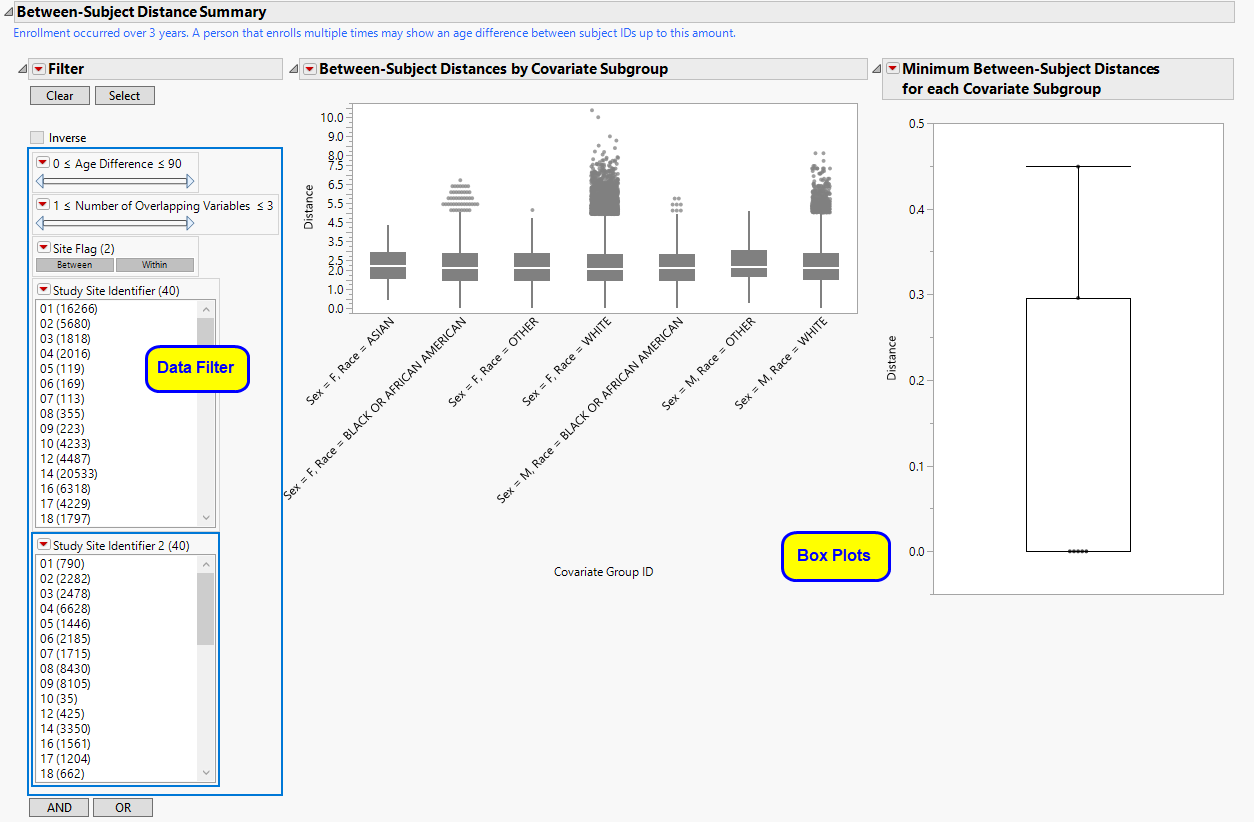
The Sex = F, Race = BLACK OR AFRICAN AMERICAN subgroup clustering section is shown below:
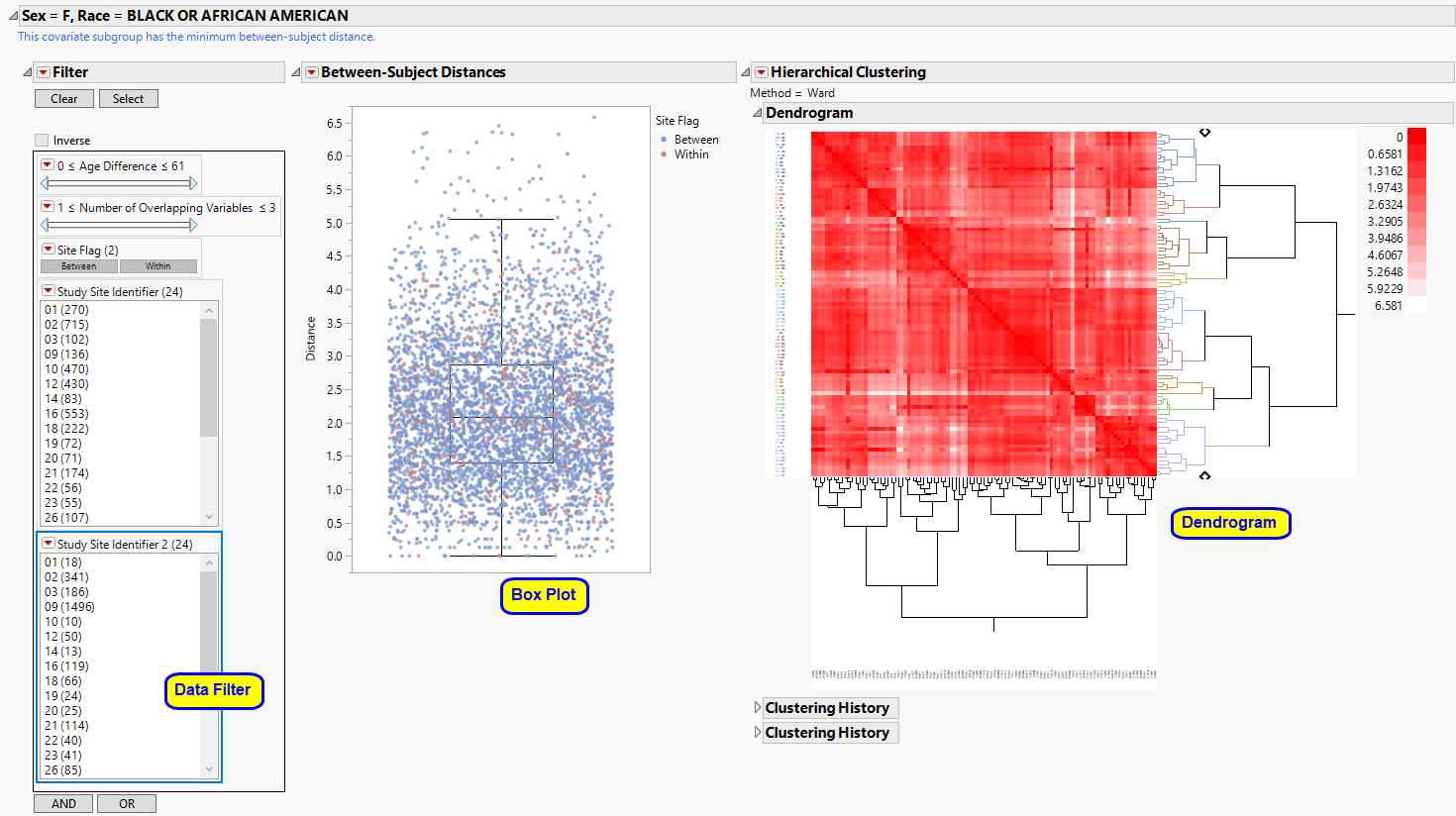
This section contains the following elements:
| • | A Box Plot showing all pairwise distances between white females across all sites. Smaller distances indicate individuals that are more similar based on pre-dose information selected for use from the dialog. Using the data filter to subset to pairs with a small age, height and weight difference, we can highlight them in the hierarchical clustering profile or examine in the data table to assess similarity. |
| • | A Dendrogram showing the Hierarchical Clustering performed to identify subsets of subjects that might be very similar, for example, a subject that has attended at least 3 sites. Points indicating highly similar pairs of subjects can be selected from the box plot, and these rows can be highlighted in the clustering heat map. |
| • | A Local Data Filter to subset histograms to data of interest, for example, to data for a particular set of study monitors. Age (and height and weight, if available) are presented to limit pairs to those that are more likely to indicate a match between two subjects. Selecting Within subsets to pairs from subjects within the same site. Selecting Between subsets to subjects from different sites. |
Action Buttons
Action buttons, provide you with an easy way to drill down into your data. The following action buttons are generated by this report:
| • | Profile Subjects: Select subjects and click  to generate the patient profiles. See Profile Subjects for additional information. to generate the patient profiles. See Profile Subjects for additional information. |
| • | Show Subjects: Select subjects and click  to open the ADSL (or DM if ADSL is unavailable) of selected subjects. to open the ADSL (or DM if ADSL is unavailable) of selected subjects. |
| • | Show Rows in Heat Map: Select points that represent pairs of subjects in the Box Plots and click  to highlight the subjects within the Heat Map and Dendrogram to see how they cluster together. to highlight the subjects within the Heat Map and Dendrogram to see how they cluster together. |
| • | Subset Clustering: On a subgroup clustering page, subsets clustering to subjects, based on pairs selected from corresponding box plot. |
| • | Revert Clustering: Click  to return a subset clustering to the original state where all subjects are clustered. to return a subset clustering to the original state where all subjects are clustered. |
General
| • | Click  to view the associated data tables. Refer to View Data for more information. to view the associated data tables. Refer to View Data for more information. |
Output includes one summary data set (named csass_sum_XXX1, by default) containing one record per subject with pre-dosing data, one data set of all pairwise distances within the covariate subgroups (named csass_alldist_XXX, by default), one data set containing minimum pairwise distances for each covariate subgroup (named csass_mindist_XXX), by default), one data set per covariate subgroup containing pairwise distances (named csass_p_Y_XXX, by default, where Y is indexed 1 to the number of covariate subgroups) and one data set per covariate subgroup containing the distance matrix of subjects within the covariate subgroup (named csass_Y_XXX, by default, where Y is indexed 1 to the number of covariate subgroups).
| • | Click  to generate a standardized pdf- or rtf-formatted report containing the plots and charts of selected sections. to generate a standardized pdf- or rtf-formatted report containing the plots and charts of selected sections. |
| • | Click  to take notes, and store them in a central location. Refer to Add Notes for more information. to take notes, and store them in a central location. Refer to Add Notes for more information. |
| • | Click  to read user-generated notes. Refer to View Notes for more information. to read user-generated notes. Refer to View Notes for more information. |
| • | Click the arrow to reopen the completed report dialog used to generate this output. |
| • | Click the gray border to the left of the Options tab to open a dynamic report navigator that lists all of the reports in the review. Refer to Report Navigator for more information. |
Methodology
No testing is performed. Subjects are clustered within each site according to the selected clustering methodology. Refer to the JMP documentation on hierarchical clustering for statistical details.
Report Options
Data and Analysis
Demographics
By default, the analysis Include Age. You can opt to Include Sex, as well, or to ignore either these if you choose.
Findings
You can opt to Include findings domains data. While all tests from all findings domains are included in the analysis by default, you can restrict the analysis to specified Findings Tests only. You can also select analysis units along with cutoff values for including events or interventions, for summarizing subgroups, and specify whether variables with missing values are allowed. You can Analyze findings using: either standard units or the original units.
Unscheduled visits can occur for a variety of reasons and can complicate analyses. By default, these are excluded from this analysis. However, by unchecking the Remove unscheduled visits box, you have the option of including them.
Interventions
You can opt to Include intervention domains data in your analysis. By default, all intervention domains are included, however, you can use the Subset of Domains to Analyze for Interventions option to restrict the analysis to specific domains.
Events
You can opt to Include event domains data in your analysis. By default, all event domains are included, however, you can use the Subset of Domains to Analyze for Events option to restrict the analysis to specific domains. Use the Include events or interventions experienced by at least this percent of patients: option to specify a minimum threshold for including an event or intervention in the analysis.
Finally, you can exclude variables missing some or any values. See Remove all variables with missing values, and Remove variables from analysis with a missing data percentage of at least: for more information.
The Summarize subgroups with at least this many subjects option enables you to generate summaries of significant subgroups of patients.
Clustering
By default, specified subjects are clustered by Sex and Race using Ward’s Hierarchical Clustering Method. Available options enable you to change both the criteria for clustering (adding Country, for example) and the clustering method used.
Filtering the Data:
Filters enable you to restrict the analysis to a specific subset of subjects based on values within variables. You can also filter based on population flags (Safety is selected by default) within the study data.
See Select saved subject filter2, Additional Filter to Include Subjects, and Subset of Visits to Analyze for Findings for more information.